msFineAnalysis AI Ver. 3
Unknown Compounds
Structure Analysis Software

A New Era of Structural Analysis of Unknown Compounds
the Fusion of Cutting-edge AI Technology and Comprehensive GCxGC
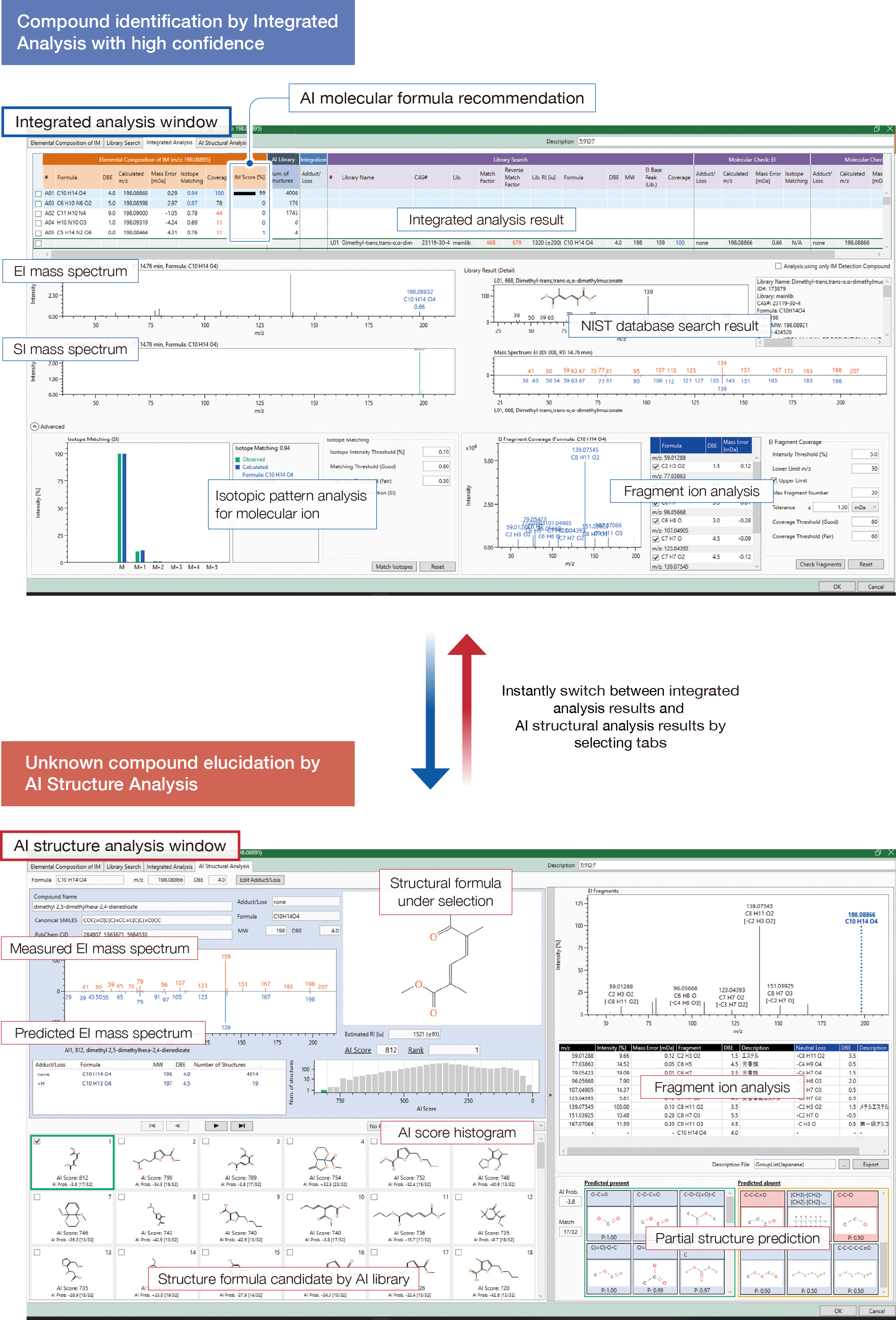
msFineAnalysis AI offers a new structure analysis tool for unknown compounds that specifically designed for the JEOL JMS-T2000GC "AccuTOF™ GC-Alpha 2.0". The "Integrated Analysis" combines GC/EI, GC/Soft Ionization high resolution data, and the "AI Structure Analysis" using four AI technologies.
Version 3 supports comprehensive two-dimensional gas chromatography (GCxGC) data analysis in addition to regular GC-MS data analysis. In complex mixtures such as petroleum samples and life science samples, the ultra-high separation of GCxGC and the structural analysis of unknown compounds using msFineAnalysis AI will prove to be very effective.
These advanced AI technologies allow msFineAnalysis AI to provide a unique automatic structure analysis capability that was not previously available for GCMS and GCxGC-MS qualitative analysis.
Features
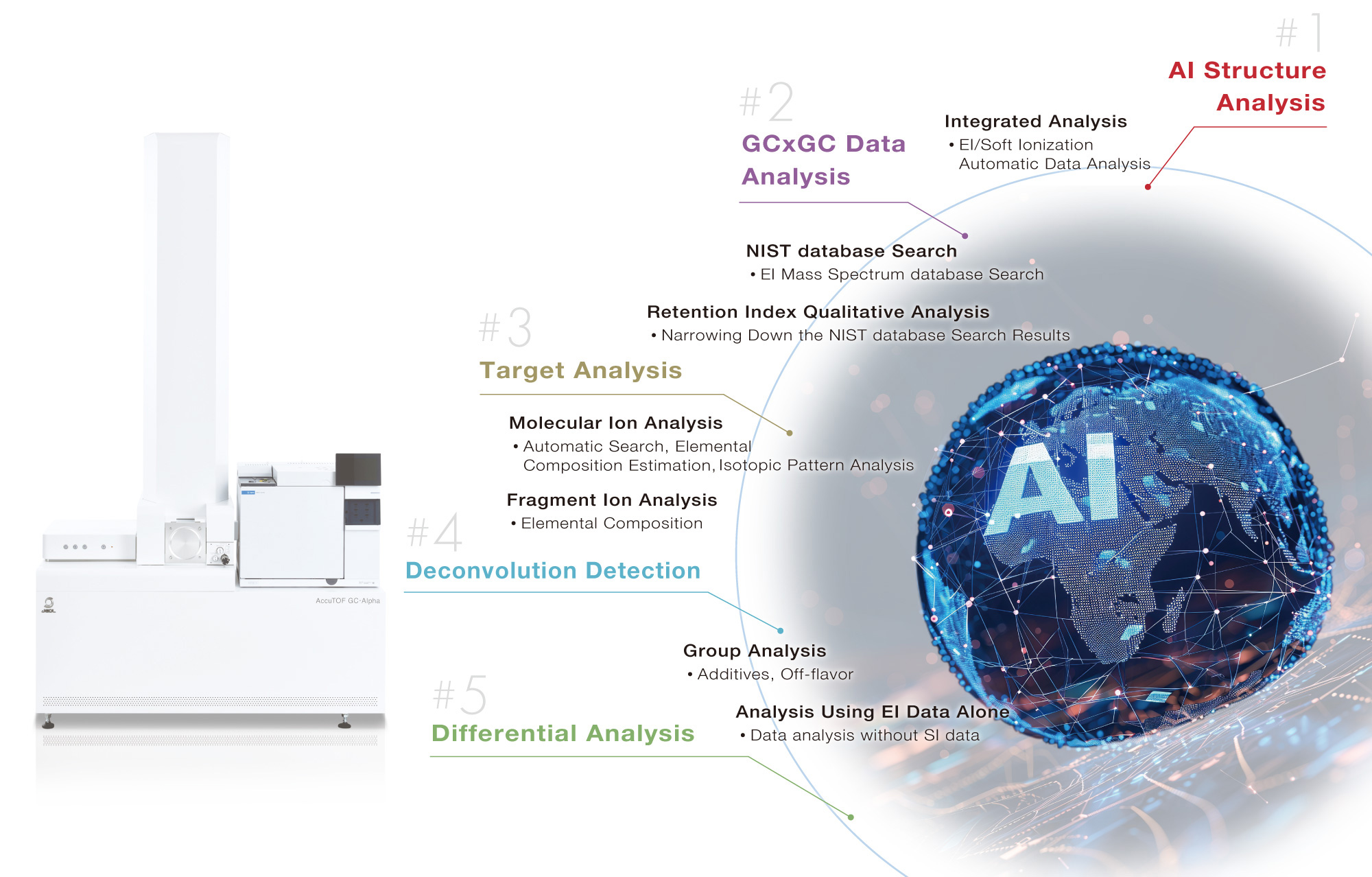
#1 AI Structure Analysis
Innovative solutions with four AI technologies
Support molecular formula determination and structural formula estimation for unknown compounds
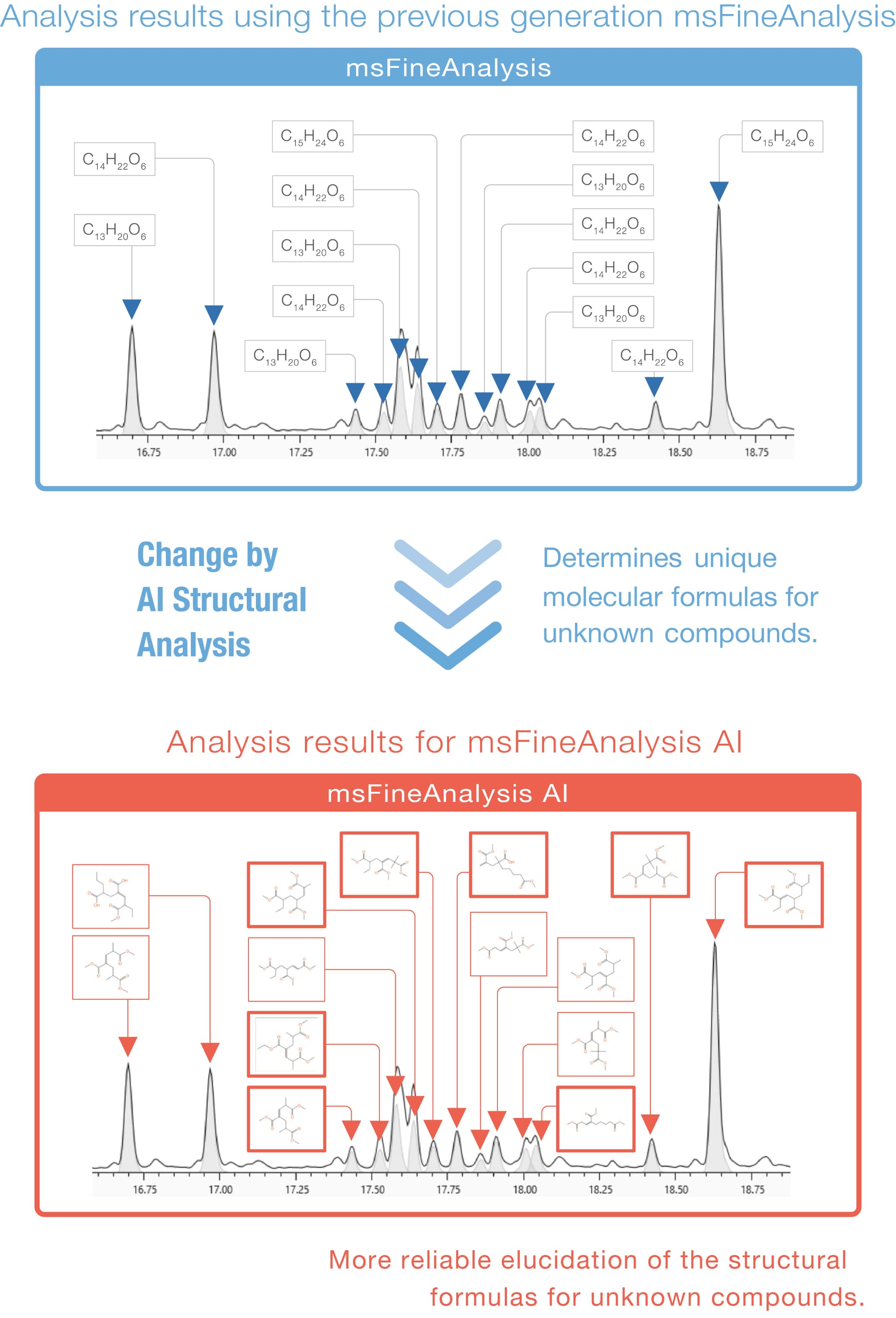
For unknown compounds (▼) not registered in the NIST database, conventional msFineAnalysis provided the molecular formula automatically. With msFineAnalysis AI, the structural formula of unknown compounds can be automatically estimated.
 The necessity for soft ionization: Reliable acquisition of molecular
formula information is the first step in structure analysis!
The necessity for soft ionization: Reliable acquisition of molecular
formula information is the first step in structure analysis!
EI mass spectral data is used for NIST database so EI methods are widely used for qualitative analysis of GC-MS samples. However, since EI is a hard ionization method, many fragment ions are observed, and in many cases, it is not uncommon to observe minimal or no signal for the molecular ions.
Additionally, for unknown compounds not registered in the NIST database, it is difficult to distinguish, using the EI mass spectra alone, whether the largest observed m/z value is actually the molecular ion or just a fragment ion. In these cases, a soft ionization method is an effective tool for determining this information.
With the AccuTOFT™ GC-Alpha, a variety of soft ionization methods including field ionization (FI), photoionization (PI), and chemical ionization (CI) are optionally available with the system. These techniques can assist in distinguishing ions (e.g., molecular ions and protonated molecules) that provide molecular weight information that makes it possible to accurately determine the molecular formula information for unknown compounds.
Since molecular formula information is an important starting point for AI structure analysis, soft ionization is critically important for identifying unknown compounds.
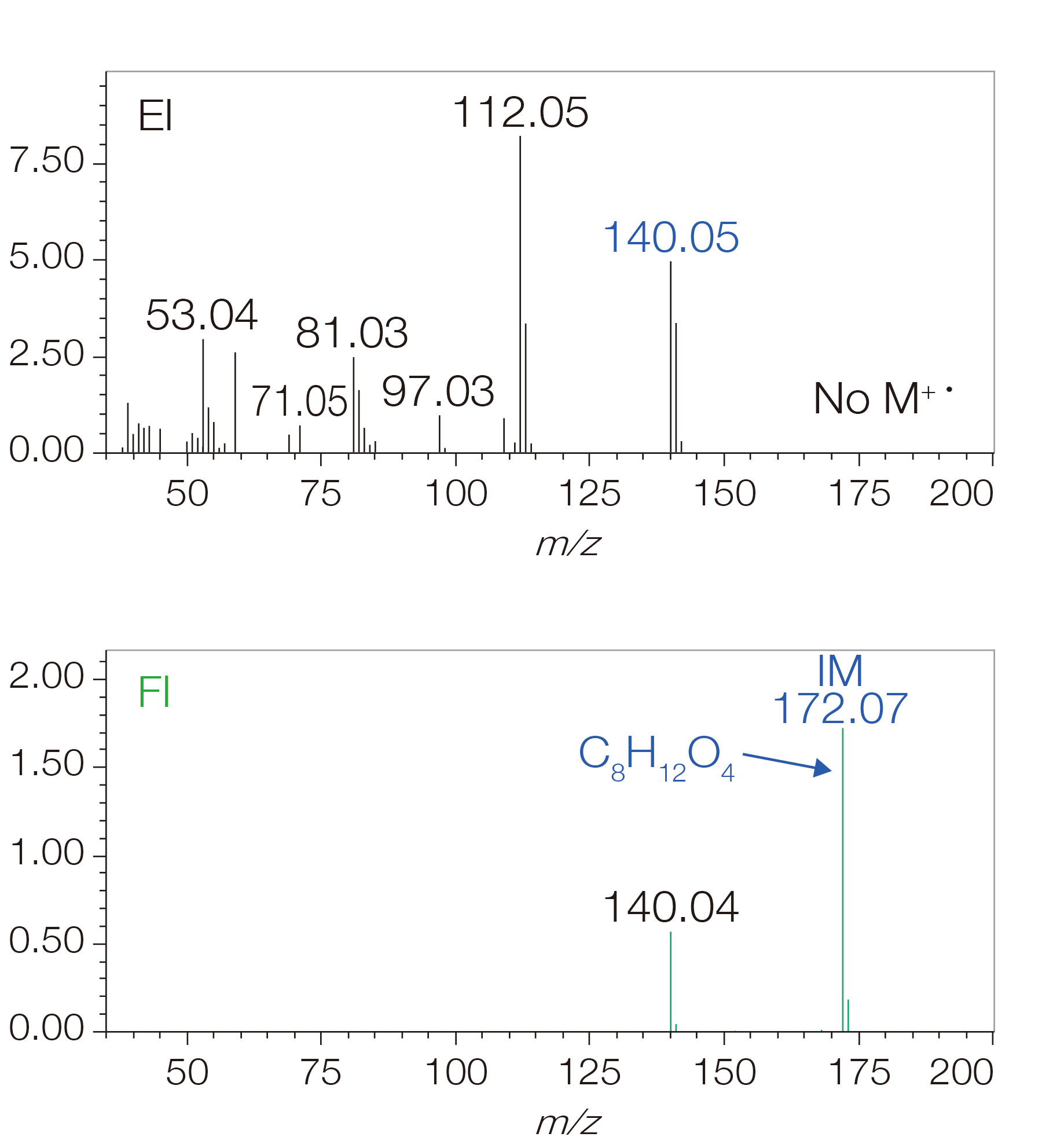
Mass spectrum of component not registered in
NIST database
 Manual Structure Analysis by Skilled Analyst vs AI Automatic Structure Analysis
Manual Structure Analysis by Skilled Analyst vs AI Automatic Structure Analysis
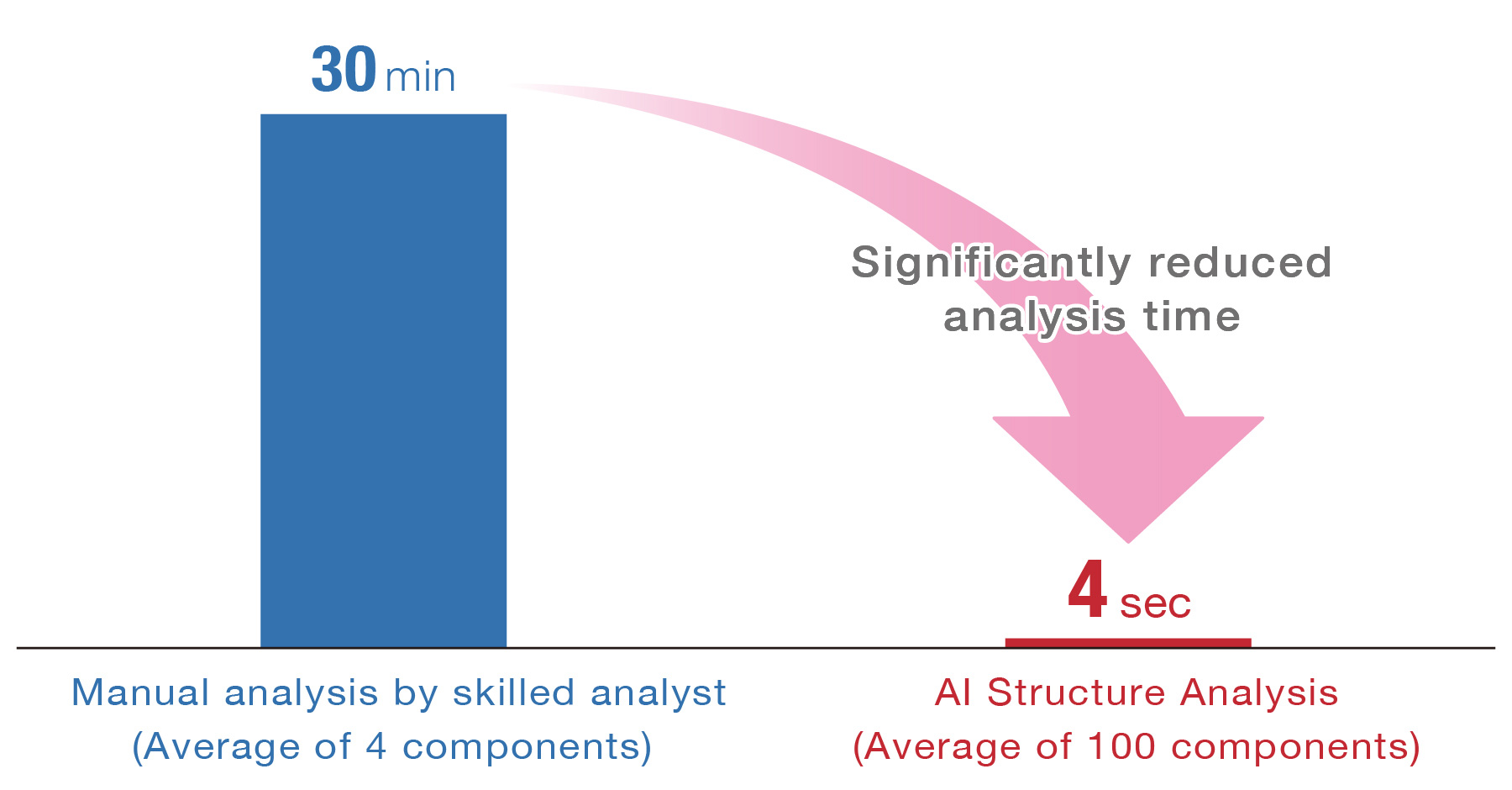
※Measured with JMS-T2000GC standard configuration PC
The time required for structure analysis was compared for the compounds observed in an acrylic resin measured by Py-GCTOFMS and were not registered in the NIST database.
Even an analyst with more than 30 years mass spectrometry experience required approximately 2 hours for structure analysis of 4 components, which is 30 minutes per component. On the other hand, AI structure analysis completed 100 components in less than 7 minutes, which is 4 seconds per component.
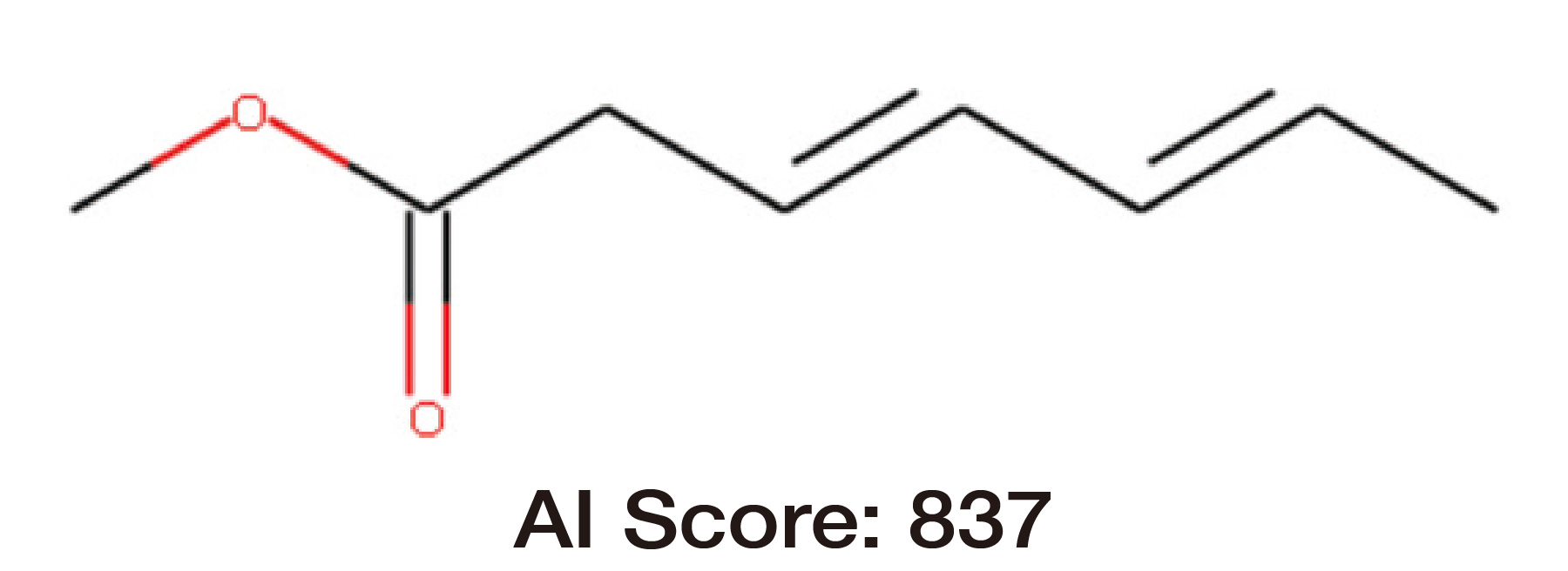
AI structure analysis score (similarity) between the structural formula estimated by a skilled analyst and the correct structural formula, indicating that the structural formula is predicted with good similarity.
Innovative solutions with four AI technologies
Automatic structure analysis without an internet connection
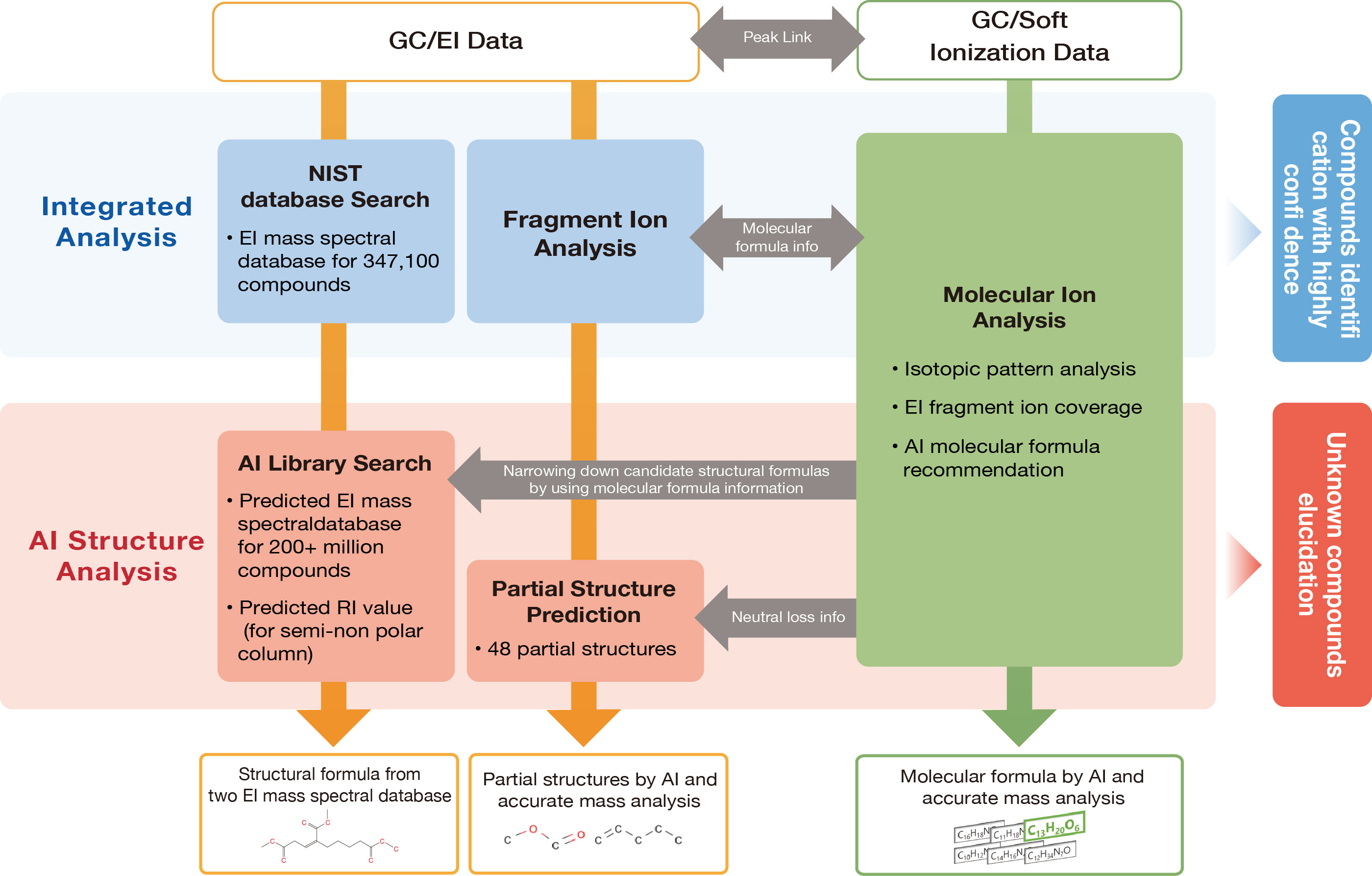
msFineAnalysis AI utilizes four different AI models (see Table 1) to automatically perform analyses on molecular formulas, substructures, and technical know-how, and provides analytical results quickly.
Table1 Four AI predictions on msFineAnalysis AI
| # | Prediction | Software function |
|---|---|---|
| 1 | EI mass spectrum | AI Library |
| 2 | Retention Index | AI Library |
| 3 | Partial structure | Present or absent for 48 partial structures |
| 4 | Molecular formula | AI molecular formula recommendation |
msFineAnalysis AI workflow
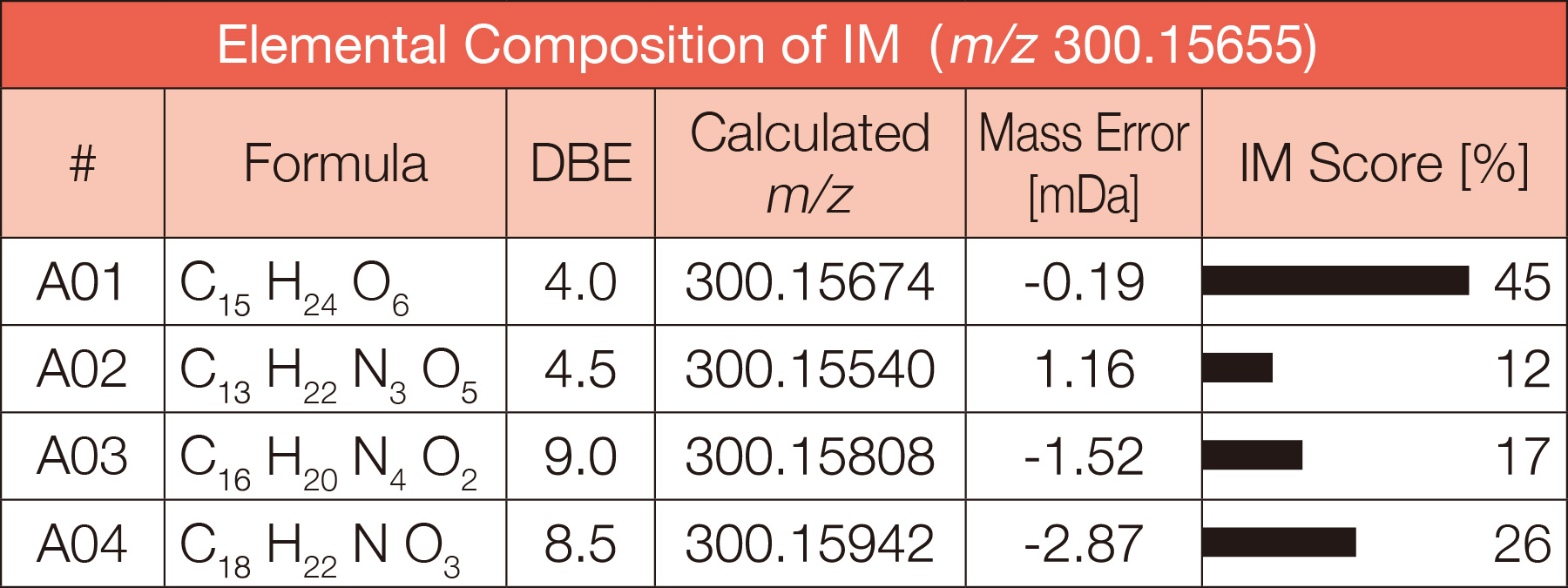
 Unique selection of molecular formulas by a fourth AI
Unique selection of molecular formulas by a fourth AI
Using a deep learning model, it assigns a score (IM score) to the candidate molecular formulas and ranks them. It is especially effective in determining molecular formulas from multiple candidates at higher mass ranges.
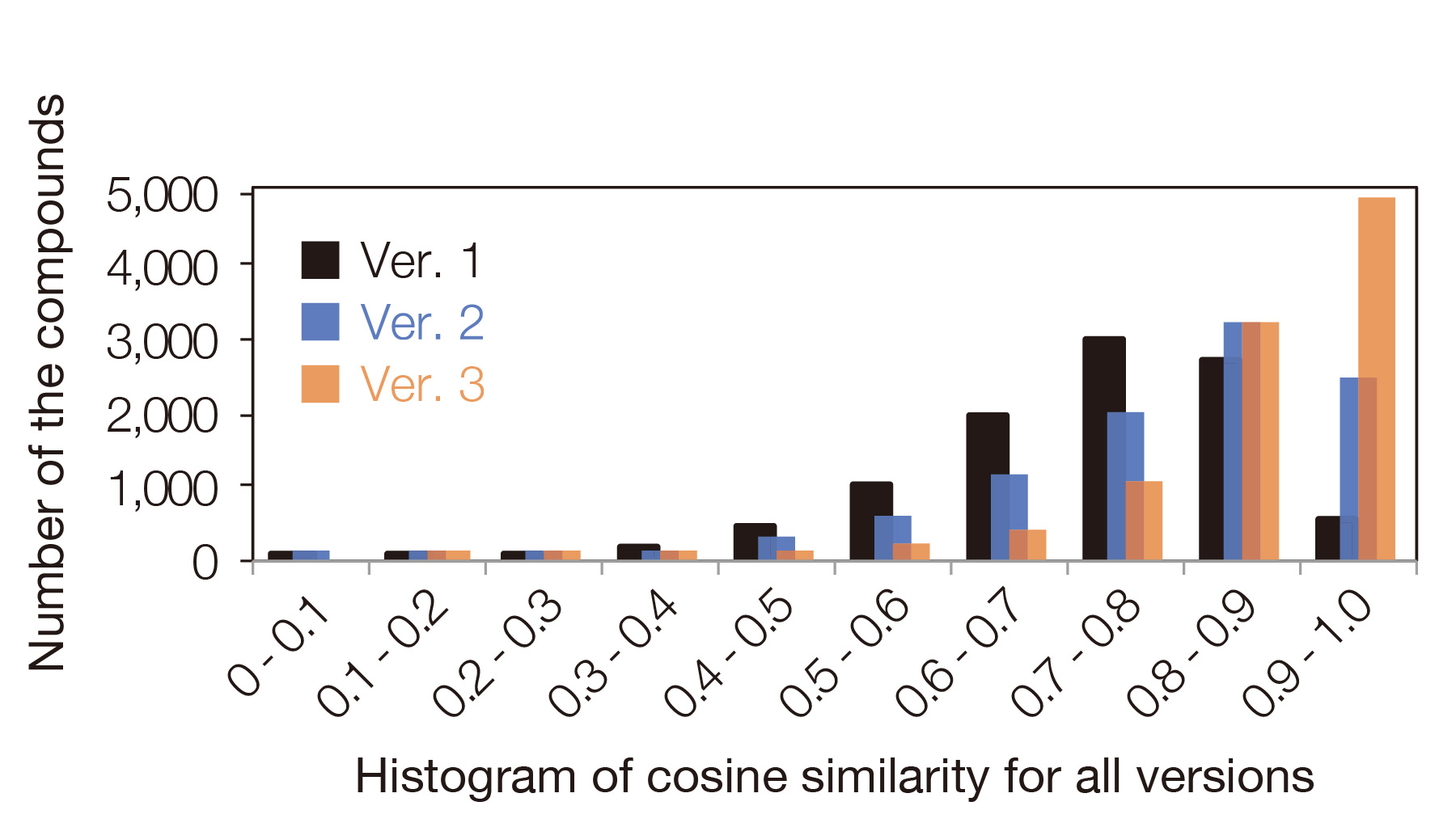
 Evolution of the AI Model
Evolution of the AI Model
The EI mass spectrum prediction model using graph convolutional networks has been further improved. Both similarity and accuracy have been greatly improved from version 1 and 2 to provide more accurate results of automated structural analysis.
Histogram of cosine similarity for all models between measured and predicted EI mass spectra of 10,000 known compounds. The AI model on version 3 has further evolved its performance, and the average value of the cosine similarity has improved to 0.86! (Ver.1: 0.72, Ver.2: 0.80)
Structural analysis of unknown compounds in foods
Flavor components in oysters were analyzed using a combination of HS-SPME GC-MS. AI structure analysis of an unknown compounds that was previously identified as 1,5-octadien-3-ol1), yielded 2,544 candidate structural formulas, which were narrowed down to 1,012 candidates using the "OH" substructure filter. The structural formula proposed by the paper was the second hit with an AI score of 936, which is quite high.
1) Kenji Ueda, Koki Yahiro, Yoshihiko Akakabe, J. Oleo Sci. 72, (7) 725-732 (2023)
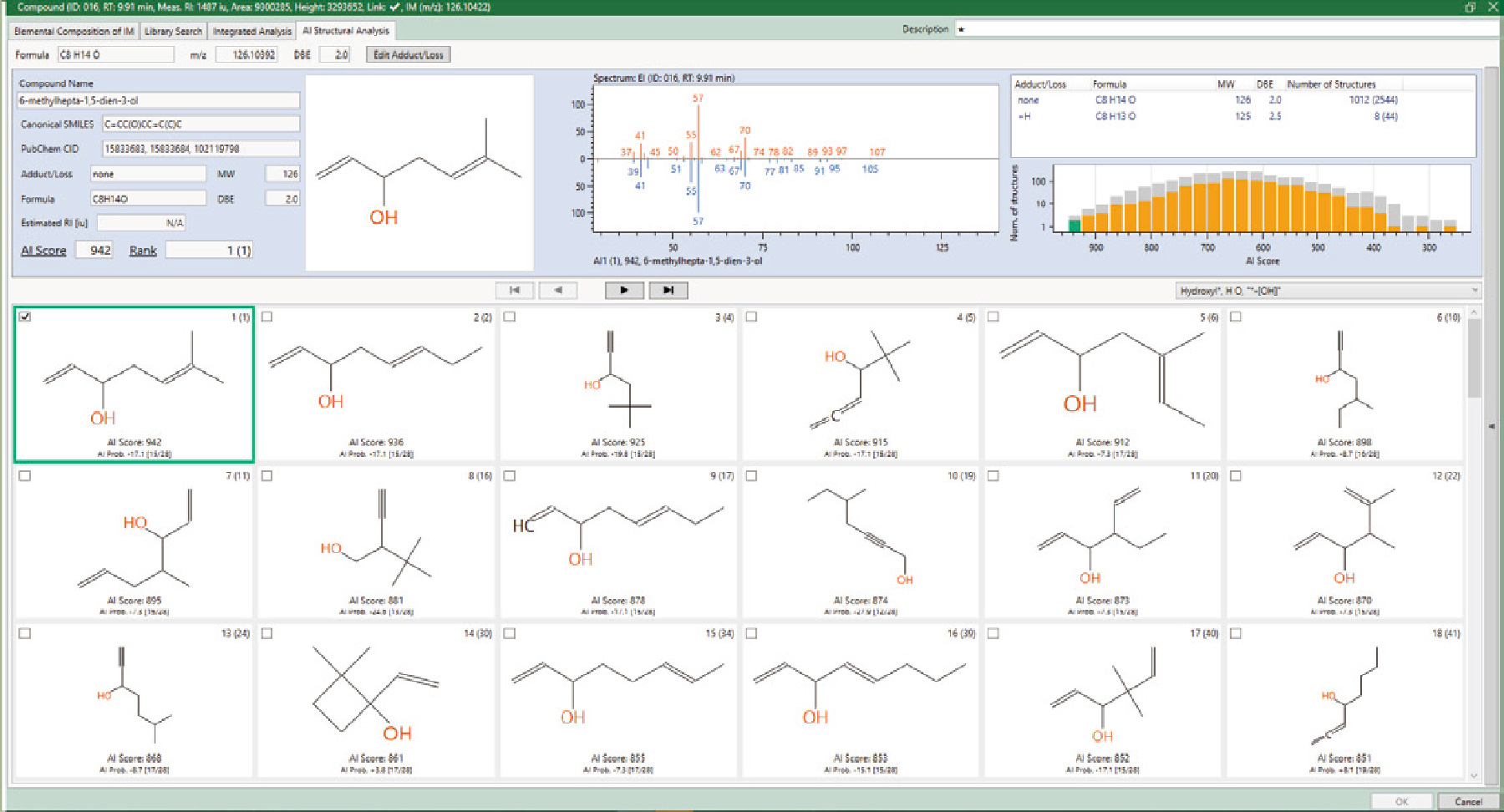
AI structural analysis result window of the flavor component
in oyster

JMS-T2000GC with a HS-SPME
(headspace – solid phase microextraction) autosampler
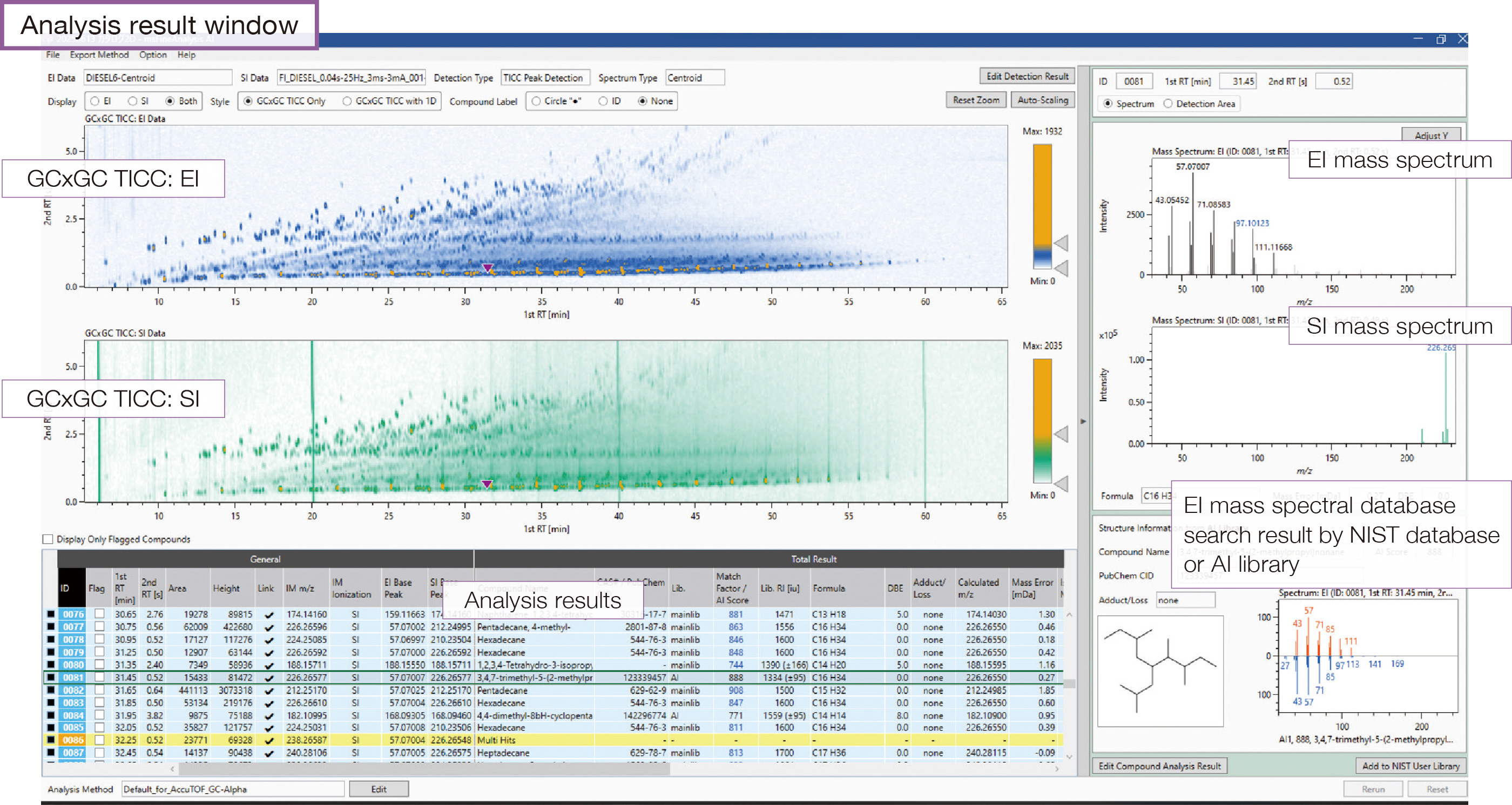
#2 GCxGC Data Analysis
Comprehensive unknown compounds analysis for complex mixture samples
GCxGC can analyze complex samples containing many compounds with high separation by using two diff erent types of columns. Ultra-high GCxGC separation chromatography combined with AI structural analysis provides a new qualitative analysis solution.
Analysis of Unknown Compounds in HeLa Cells
Water-soluble metabolites in HeLa cells were derivatized with TMS and measured by GCxGC-TOFMS, and a total of 674 metabolites were detected.
In a previous study2), an unknown compound identified as N-methyluridine monophosphate (N-methyl UMP)—a substance not registered in the NIST database—was subjected to AI-based structural analysis. As a result, N-methyl UMP was obtained as the top candidate, with a good AI score of 805. In addition, N-methyl UMP and nucleic acid-related compounds have similar retention times in the 2nd column, indicating that they are compounds with similar polarity.
2) Lai Z., Tsugawa H., et al. (2018). Nature Methods, 15 (1), 53–56. DOI: 10.1038/nmeth.4512
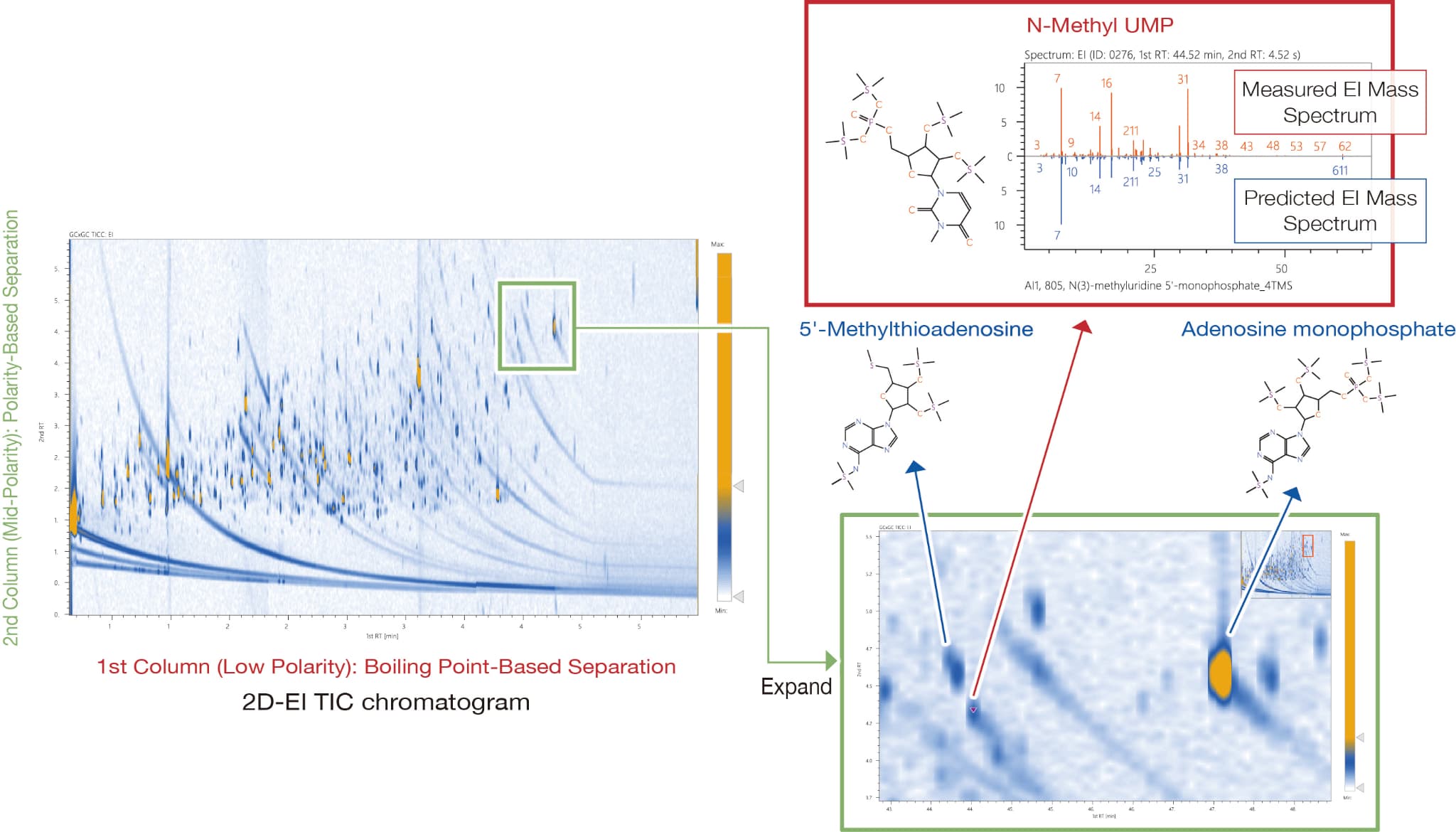
This sample was kindly provided by Professor Hiroshi Tsugawa of Tokyo University of Agriculture and Technology.
Detailed Analysis of Pyrolysis Oil
Pyrolysis oil generated from a mixture of polyethylene (PE), polypropylene (PP), and polyvinyl chloride (PVC) was analyzed by GCxGC-TOFMS. As a result, it was possible to clearly separate alkanes and alkenes derived from PE/PP, which are difficult to separate by 1D GC. In addition, the molecular formulas of the peaks were identified using the soft ionization method. For unknown compounds that were not registered in the NIST database, the structural formulas were obtained using AI structural analysis.
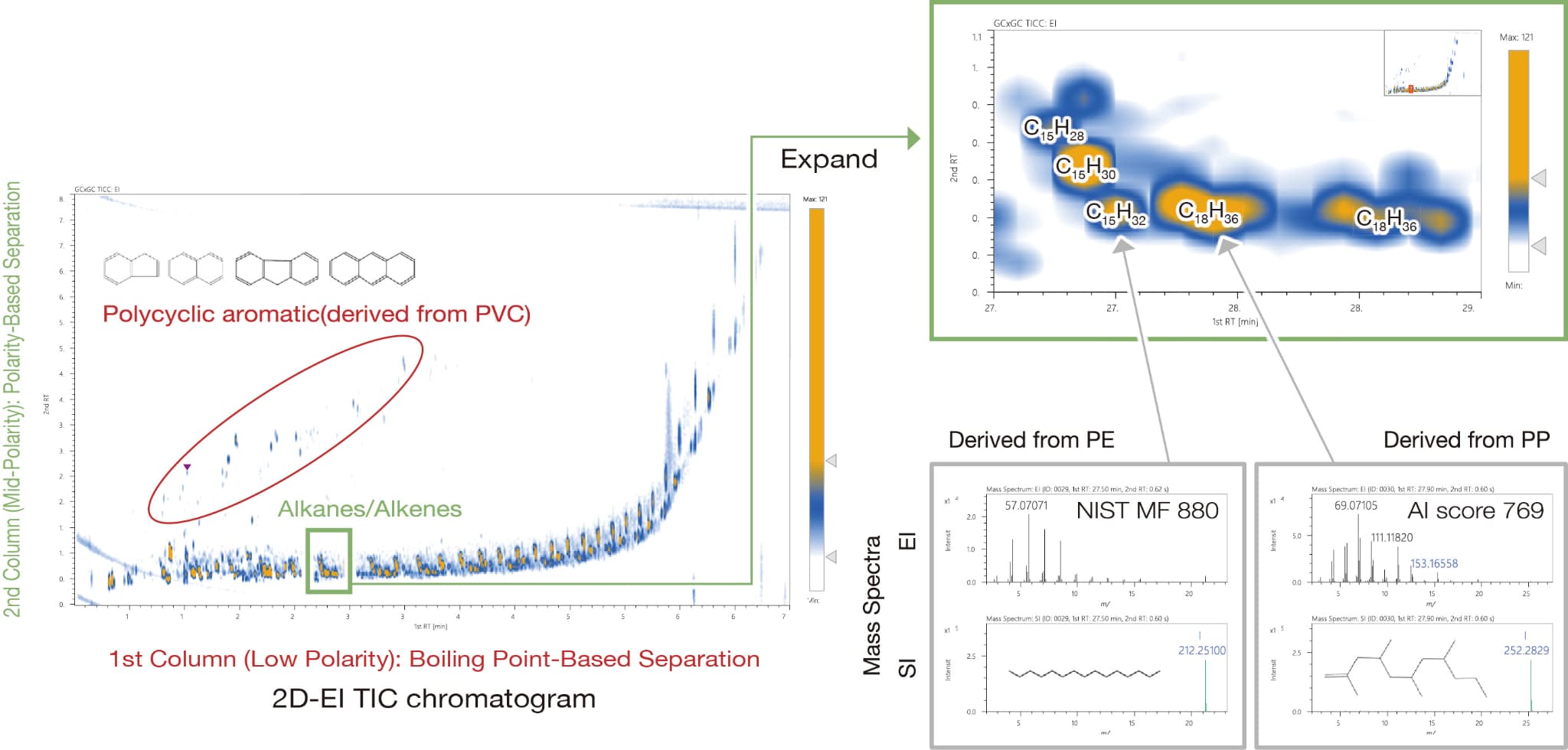
This sample was kindly provided by Associate Professor Shogo Kumagai of Tohoku University.
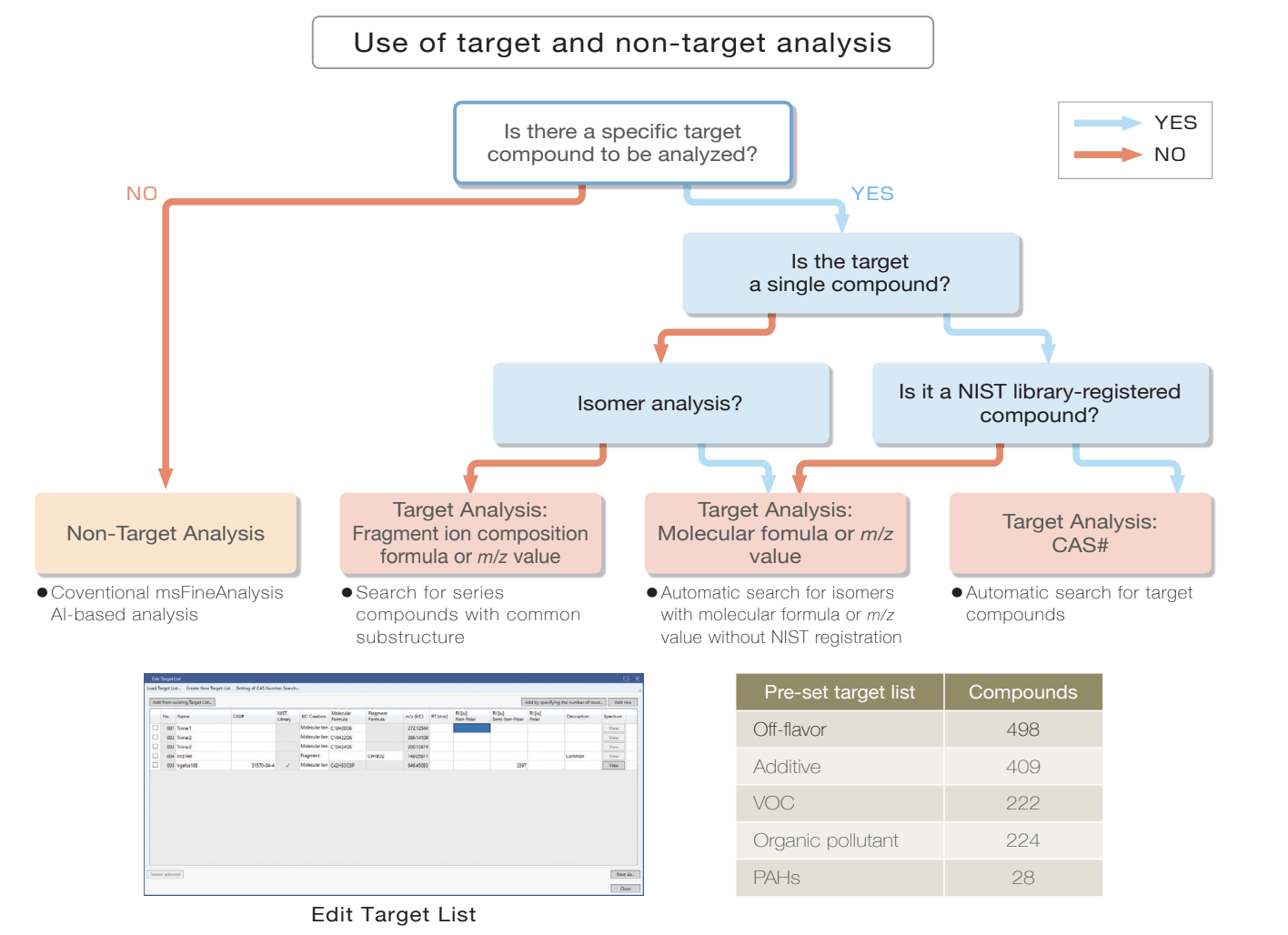
#3 Target Analysis
Rapid search for known compounds
The target analysis function automatically searches for compounds based on compositional formula, m/z value, and CAS#.
Preset target analysis lists are available. Additionally, user defined lists can be created and customized for the samples and their analytes of interest. Integrated analysis and AI structural analysis of compounds detected by target analysis are also available.
Target analysis of flavor and off-flavor components in foods
msFineAnalysis AI supports not only non-target analysis but also target analysis. It automatically searches for target compounds based on compositional formula, m/z value and CAS#.
For the data of flavor components in lemon juice, 10 compounds were extracted when analyzed with a target list of 498 off-flavor components. The result window on the lower right shows the detailed analysis result of Citral among the 10 components.
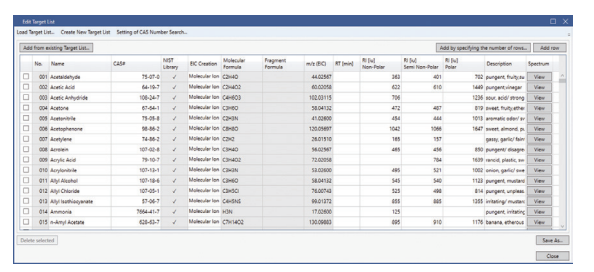
Target list of 498 off-flavor components
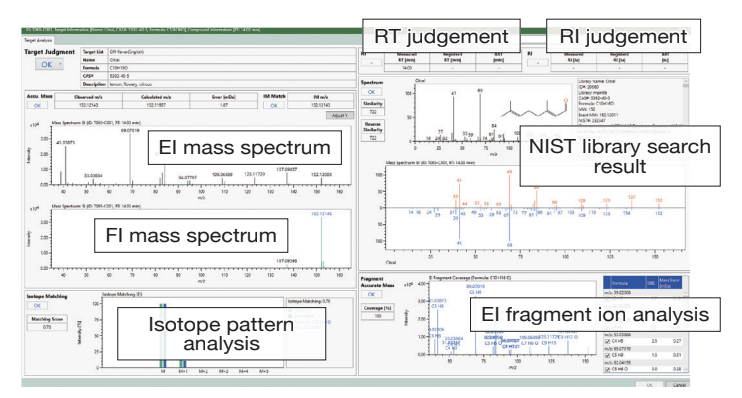
Detailed analysis of Citral present in lemon juice
#4 Deconvolution Detection
Chromatographic peak deconvolution can detect trace components that may not be obvious in the TICC due to the coelution of several components.
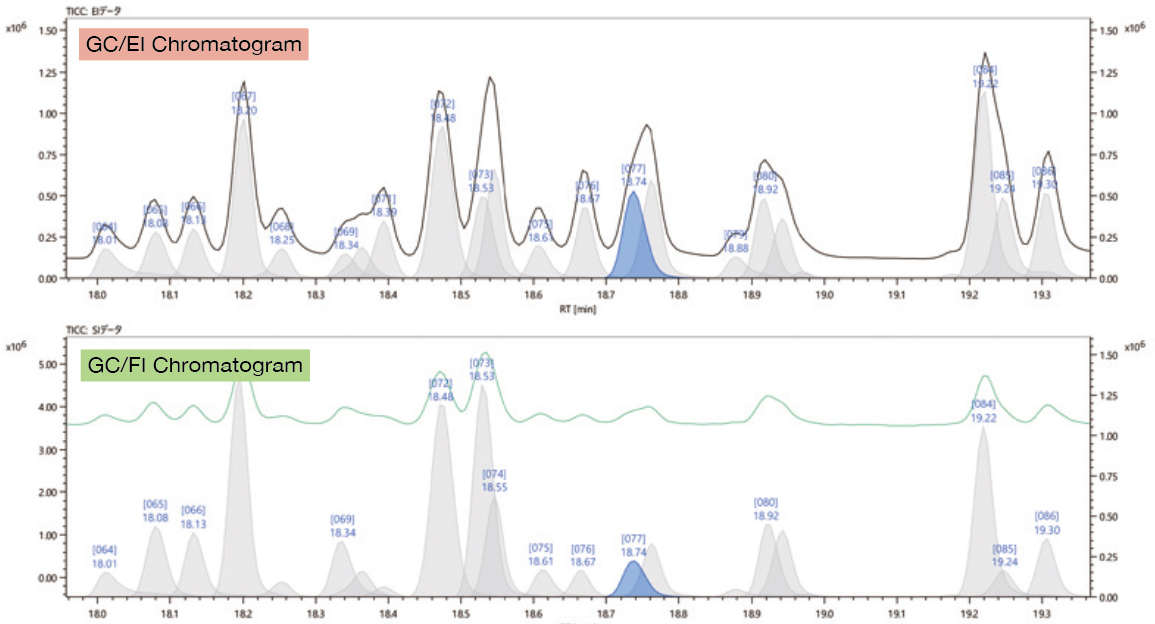
EI: black solid line: TICC, gray peaks: deconvolution peak (blue: currently selected)
FI: green solid line: TICC, gray peaks: deconvolution peak (blue: currently selected)
This step simplifies the data analysis process by defining which ions go with each compound and eliminates the need for creating extracted ion chromatograms (EICs).
#5 Two Sample Comparison (Differential Analysis)
This function uses a volcano plot, with the p-value on the vertical axis representing reproducibility, and the intensity ratio between two samples on the horizontal axis.
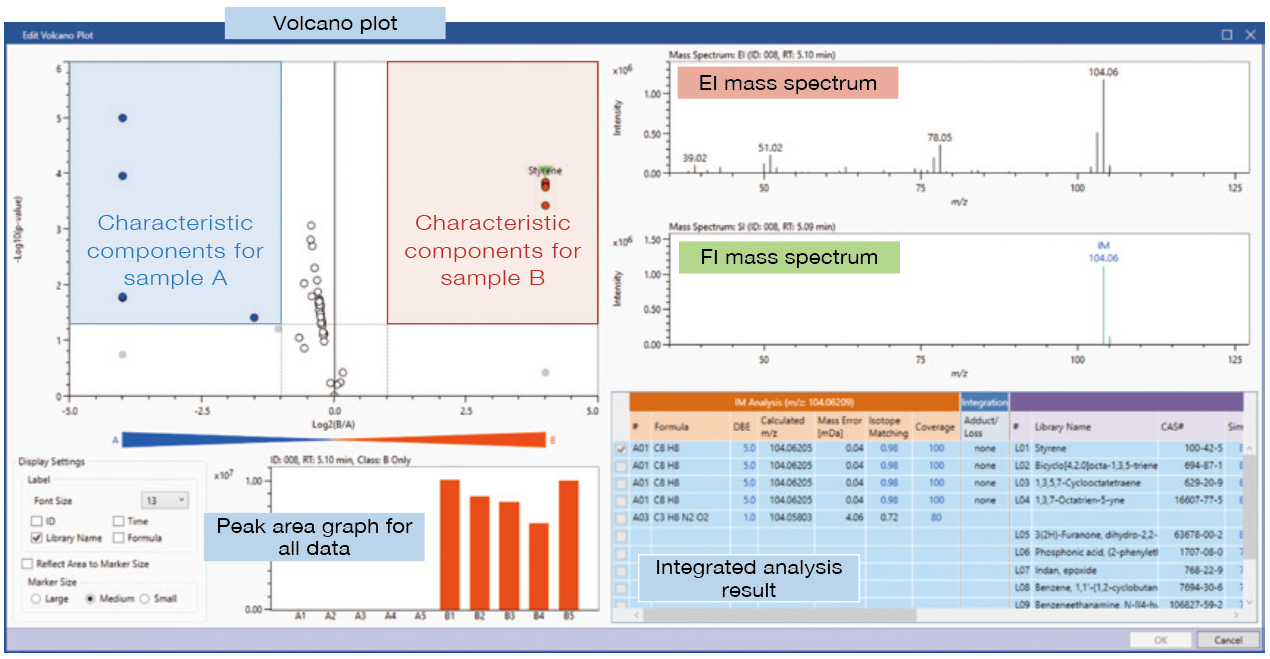
Detailed analysis – Volcano plot
(A: Reference product, B: Defective product)
This information enables a visual confirmation of the differing components between two samples. For example, it is possible to confirm if a component increases or decreases when comparing a reference product to a defective product or to identify characteristic components in a new material by comparing it to an existing material. For a two sample comparison, it is possible to set n=1, 3, 5 for the number of measurements for each sample.
Specifications
Major Specifications
JMS-T2000GC AccuTOFTM GC-Alpha 2.0
| Mass Resolution | 30,000 @ m/z 614 |
|---|---|
| Mass Accuracy | 1 ppm @ EI standard ion source |
| Ionization Methods | EI、CI、PI、FI、FD、DEI、DCI |
msFineAnalysis AI Ver. 2
| Specifications |
*1【Number of compounds registered in AI Library】
|
|---|
Catalogue Download
msFineAnalysis AI Unknown Compounds Structure Analysis SoftwareAutomatic
Application
GC-TOFMS Application: Qualitative Analysis of Chemical Components in an Herbal Medicine
Related Products

JMS-T2000GC AccuTOF™ GC-Alpha 2.0 Gas Chromatograph - Time-of-Flight Mass Spectrometer
The ultimate GC-MS system that pursues the ultimate in performance and functionality, the JMS-T2000GC AccuTOF™ GC-Alpha, has evolved into version “2.0” with the integration of a fully automated standard sample inlet and msFineAnalysis AI.
More Info
Are you a medical professional or personnel engaged in medical care?
No
Please be reminded that these pages are not intended to provide the general public with information about the products.
