single particle analysis
single particle analysis
An electron microscopy method to obtain the three-dimensional (3D) structure of biological macromolecules, such as proteins and nucleic acids, by analyzing electron microscope images of the macromolecules (proteins, etc.) recorded as dispersed particles (single particles). A solution containing a protein is made into a thin film on a holey carbon film attached on an EM grid and is then rapidly frozen so that the particles are embedded in an amorphous ice film. TEM images are taken at Liq.N2 temperature, and the 3D structure is reconstructed by image processing and analysis.
A number of projection images of the 3D structures of protein particles in various orientations are recorded in the acquired Cryo-TEM images. Each particle image is rotated and translated in the image plane to collect the images having the same external shape and the same density distribution to sort them into groups of different 3D orientations. Then, the images in the same groups are added and averaged to improve the signal-to-noise ratio of the images. Next, the Euler angles of the projection orientations (angles of the particles against the image plane in the present case) are estimated. Finally, the 3D structure of the protein is reconstructed by back projection of these particle images along the estimated Euler angles.
Single particle analysis has been dramatically improved in its achievable resolution and practical application by the advent of a CMOS direct electron detector, together with innovations in specimen preparation techniques and analysis software programs. The method is applicable to an extremely small amount of sample solution (protein concentration: several mg/mL, solution volume: several µL/grid), and is especially effective for structural analysis of proteins that are difficult to crystallize. As of 2018, the highest resolution achieved with the method is 0.16 nm.
Specimen preparation and observation method
A highly purified protein solution is prepared in which protein particles with a homogeneous structure are dispersed. The protein particles are ice-embedded by rapid freezing (at –180 °C or less) so as to preserve their structures. It is important that the ice film embedding the protein particles must be prepared as thin as possible to produce high contrast TEM images because proteins are composed of light elements (C, H, O, N, S, etc.) and produce low contrast against the ice film by a small difference between their densities.
The specimen grid is inserted into the microscope column with its frozen state kept. TEM images of the specimen are recorded at Liq.N2 temperature. To suppress the damage to the specimen due to electron-beam irradiation, the electron dose onto the specimen must be set low. The specimen is observed using phase contrast because protein particles consisting of light elements are difficult to observe using scattering contrast. For observation of protein particles, a large amount of defocus of the objective lens (–0.5 to several µm) is used to enhance the image contrast, particularly in a low spatial frequency range to allow the detection and orientation determination of the particles.
Image acquisition
Images of protein particles recorded at a low electron dose have a very low-signal-to-noise ratio (S/N). To obtain the particle images with a high S/N, hundreds to thousands of Cryo-TEM images are collected, from which several tens of thousands to several hundreds of thousands of particle images are extracted and added and averaged after image classification and alignment. In recent years, several thousands of Cryo-TEM images are automatically acquired over several days using an automatic image acquisition system.
It is emphasized that the electron-beam-induced drift of ice-embedded protein particles is the main cause of degrading the resolution of the TEM image in the single particle analysis method. Extremely high frame rate of direct electron detectors (scintillator not used) that allow single electron counting and movie-mode image recording has improved the resolution of the TEM image. Recording a TEM image in the movie mode at a frame rate of 5 to 10 frames/sec allows the particle image drift to be greatly reduced by post image processing, namely motion correction of the particle images between the frames enables acquisition of high-resolution protein particle images.
Image analysis
From several thousands of the Cryo-TEM images, several tens of thousands to several hundreds of thousands of the protein particle images are automatically extracted using a particle detection software program. Since the particle images are projected in various orientations, the extracted images are rotated and translated so as to sort them out into many classes of the projection images in the same orientations. Then, the images sorted in each class are aligned in their positions and orientations and are averaged. As a result, 100 to 200 protein particle images with a high S/N are obtained. In this step, the 3D orientations of the particle images (Euler angles or the angles of the particles against the image plane in the present case) are not yet determined.
To obtain the Euler angles, first an initial particle model with arbitrary orientations is given. Secondary, the projection images of the model are produced. The Euler angle of each particle image is estimated by a comparison of the projection images and the TEM image. The particle TEM images with the estimated Euler angles are subjected to back projection to produce an initial 3D structure of the particle. Then, this 3D structure is subjected to projection again to create the projection images. The re-projected images and the particle TEM images are compared to estimate the Euler angles again. These particle images are back-projected to obtain a better 3D structure in the next step. These steps are repeated until the obtained 3D structure is converged. In each step of calculations, the results are evaluated by statistical approaches. Finally, the most probable 3D structure is obtained.
Comparison with other analytical methods
Structural analysis methods using electron microscopy include Electron Crystallography, Electron Tomography and Single Particle Analysis. In Electron Crystallography, a 2D or thin 3D crystal is prepared and analyzed using techniques similar to those of X-ray Crystallography. Electron Crystallography provides a resolution better than 0.2 nm. In Electron Tomography, one specimen is placed in the specimen holder in the microscope column, and is subjected to serial tilt-series imaging at various angles. Then, back projection is applied to the acquired projection images for obtaining the 3D structure. In the method, it is not possible to improve the signal-to-noise ratio and the resolution of the images by averaging. For biological applications, the method is used for analysis of functional structures at the cell level, its resolution being on the order of several nm to 10 nm. But when the tomogram contains many particles of the same proteins, the use of the sub-tomogram averaging method improves the resolution close to 0.3 nm.
Conventionally-used structural analysis methods of proteins are X-ray Crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy. The former method requires a crystal with a size of 10 µm or more. Thus, the method cannot be applied to proteins that are unable to crystallize. The latter method can be used only for proteins with a molecular weight of ~50,000 or less. However, Single Particle Analysis does not have such restrictions and its resolution has been becoming as high as that of X-ray Crystallography, although at present it is still difficult to apply this method to proteins with molecular weight less than 100,000 because of low S/N of their TEM images. In the future, the advancement and the more use of Single Particle Analysis are highly expected.
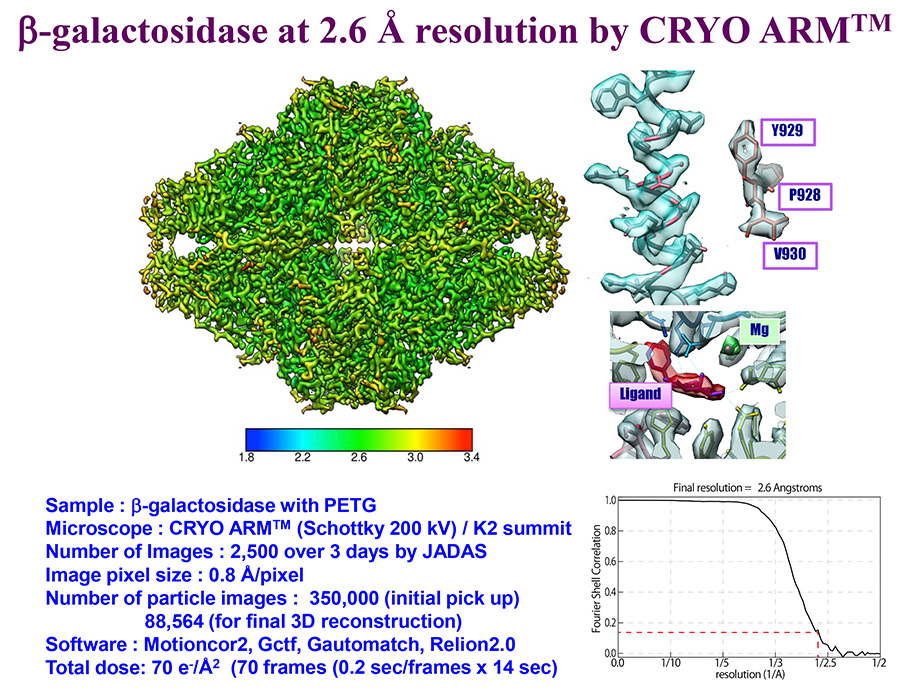
Data courtesy: Specially Appointed Professor Keichi Namba and Specially Appointed Associate Professor Takayuki Kato, Osaka University
Related Term(s)
Term(s) with "single particle analysis" in the description
Are you a medical professional or personnel engaged in medical care?
No
Please be reminded that these pages are not intended to provide the general public with information about the products.