Analysis of degraded polymethyl methacrylate by UV irradiation using high-resolution MALDI-TOFMS and pyrolysis-GC-QMS
MSTips No.324
Polymers can be degraded by the effects of light, oxygen, heat, etc. so it is important to understand how the polymer structures change during degradation. Pyrolysis gas chromatograph quadrupole mass spectrometer (Py-GC-QMS) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOFMS) are powerful tools for analyzing polymeric materials.
Py-GC-QMS is a method that instantaneously heats a sample with a pyrolyzer and then analyzes the pyrolysis products by GC-MS. Since most of the pyrolysis products are related to monomers and dimers, this technique allows for easy identification of the polymer substructures which is useful for identifying changes to the polymer when degradation occurs. MALDI-TOFMS involves a soft ionization technique that can directly ionize and analyze the intact polymer molecules and often produces singly-charged ions even for high molecular weight compounds.
As a result, the m/z axis of the mass spectrum is equal to the mass of the ions, thus making it easy to interpret polymer distributions. Additionally, when MALDI is used with a high-resolution TOFMS, the accurate mass of each ion in the polymer series can be used to calculate their elemental compositions. Moreover, the molecular weight distribution of polymers can be calculated from the ion intensity distribution. In this work, we used Py-GC-QMS and high-resolution MALDI-TOFMS to evaluate the effects of UV irradiation on polymethyl methacrylate (PMMA).
Experiment
Polymethyl methacrylate (molecular weight 5000, end groups H/H) purchased from Agilent Technologies, Inc. was used for the sample. For the degraded sample, the PMMA was UV-irradiated for 0.5 hours using a Portable Cure 100 (SEN LIGHTS Co., Ltd.).
A JMS-Q1500GC equipped with a pyrolyzer (Frontier Labs) was used for the Py-GC-MS measurements. For these measurements, the samples before and after UV irradiation were weighed to about 0.2mg and measured using the conditions shown in Table 1. AnalyzerPro (SpectralWorks Ltd.) was used to directly compare their differences, and NIST library searches were used to identify the compounds that showed significant differences between the before and after UV irradiation.
A JMS-S3000 was used for the MALDI-TOFMS measurements. The samples before and after UV irradiation was dissolved in tetrahydrofuran (THF) at 1 mg / mL. DCTB (20 mg / mL THF solution) and sodium trifluoroacetate (NaTFA, 1 mg / mL THF solution) were used as matrix and the cationization agent, respectively. The mixture of the sample solution, matrix solution and cationizing agent solution at a ratio of 1:10:1 (v/v/v) was pipetted on the target plate and air-dried. Mass spectra were acquired in high resolution SpiralTOF positive ion mode. Afterwards, Kendrick mass defect (KMD) analysis was performed by using the JEOL msRepeatFinder software.
Table 1 Measurement condition of Py-GC-QMS
| Pyrolysis condition | |
|---|---|
| Pyrolyzer | PY-3030D (Frontier Laboratories Ltd.) |
| Pyrolysis temperature | 600°C |
| GC condition | |
|---|---|
| GC | 7890A (Agilent Technologies, Inc.) |
| Column | ZB-5 (Phenomenex Inc.) 30 m × 0.25 mm I.D., df=0.25 µm |
| Injection port temperature | 320°C |
| Oven temperature | 40°C (2 min) →20°C / min→320°C (20min) |
| Injection mode | Split 100:1 |
| Carrier gas | He, 1.0 mL / min (Constant Flow) |
| MS condition | |
|---|---|
| Spectrometer | JMS-Q1500GC (JEOL Ltd.) |
| Ion source temp. | 250°C |
| Interface temp. | 320°C |
| Ionization mode | EI |
| Ionization energy | 70 eV |
| Ionization current | 50 µA |
| Mesurement mode | Scan (m/z 29 to 600) |
| Relative EM Voltage | 100 V |
Results of Py-GC-MS
The total ion current chromatogram (TICC) before and after UV irradiation is shown in Figure 1. The major components observed in TICC were monomer, dimer, and trimer, with no significant differences before and after UV irradiation.
AnalyzerPro was then used to analyze the differences and found the peaks [1]-[4] at R.T. 9.00 to 10.00 min (Figure 2A) observed only after UV irradiation. Next, a NIST library search was performed for the peak [1]. The highest similarity (M.F.) was 715 (Figure 2B), which was not valid considering the structure of PMMA; however, the fifth candidate compound (Figure 2C, M.F. of 687) was close to the partial structure of PMMA.
The mass spectrum of the peak [1] is shown in Figure 3A. Considering the results of Py-GC-QMS and MALDI-TOFMS on the next page, the estimated structure is shown in Figure 3B.
In the results of Py-GC-QMS, a clear difference was observed before and after UV irradiation; however, the library search can't reveal the partial structure of the polymer by pyrolysis.
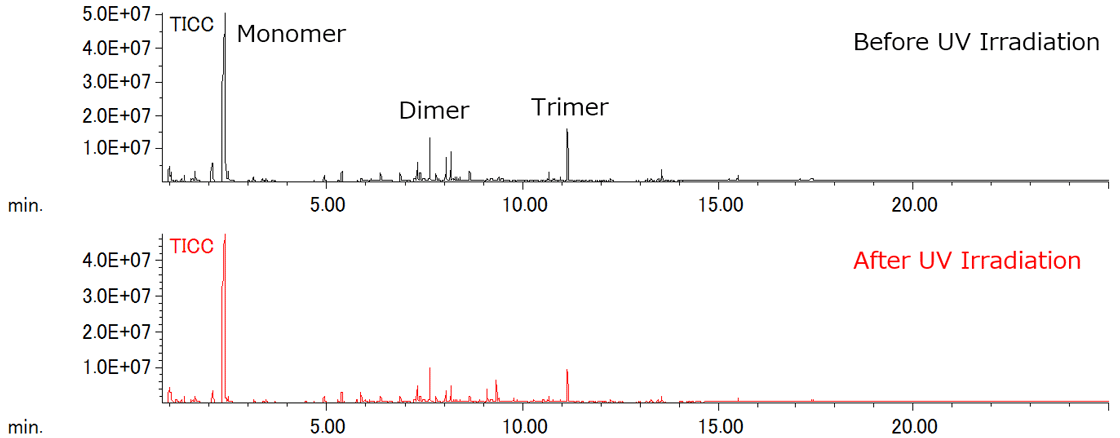
Figure 1 TICCs of PMMA before and after UV irradiation using Py-GC-QMS
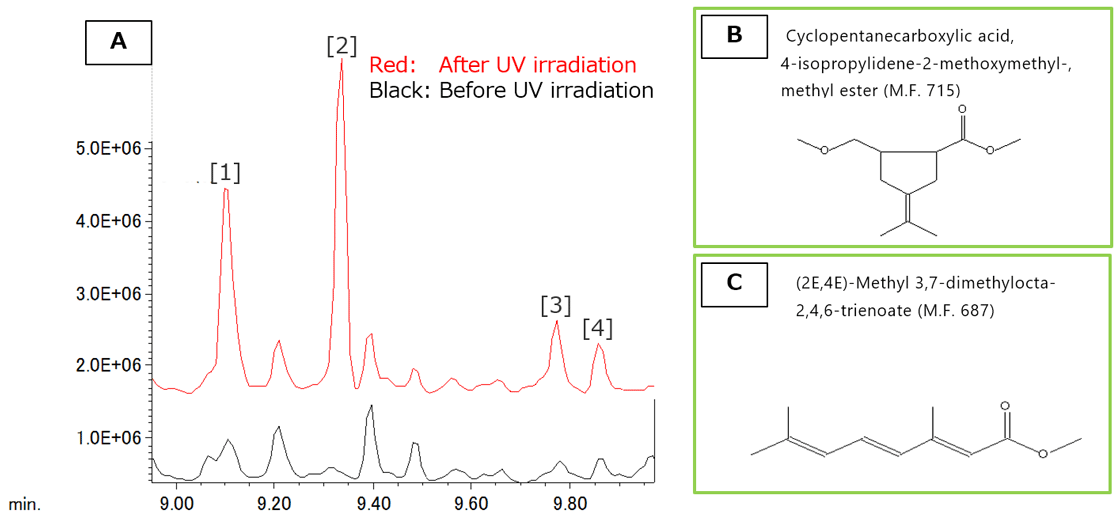
Figure 2 Difference of TICC before and after UV irradiation
![Fig. 3 Mass spectrum of Peak [1] in Figure 2A](./product_file/file/2027_05_en.png)
Figure 3 Mass spectrum of Peak [1] in Figure 2A
Results of MALDI-TOFMS
The MALDI-TOFMS mass spectra before and after UV irradiation are shown in Figure 4. Sodium adduct PMMA ions ([M+Na]+) with the H/H end groups were observed in the mass spectra for both before and after UV irradiation.
Figure 4b shows enlarged mass spectra around m/z 3700; a peak estimated as a few C2H4O2 (60.020 u) losses were observed in mass spectra after UV irradiation. The Kendrick mass defect (KMD) plots for both mass spectra are shown in Figure 5.
It clearly showed one to three C2H4O2 losses from PMMA due to photodegradation. Furthermore, the molecular weight became lower after UV irradiation. Considering the results of MALDI-TOFMS and Py-GC-QMS, it is suggested that the main chain structure changes as shown in Figure 6 by UV irradiation.
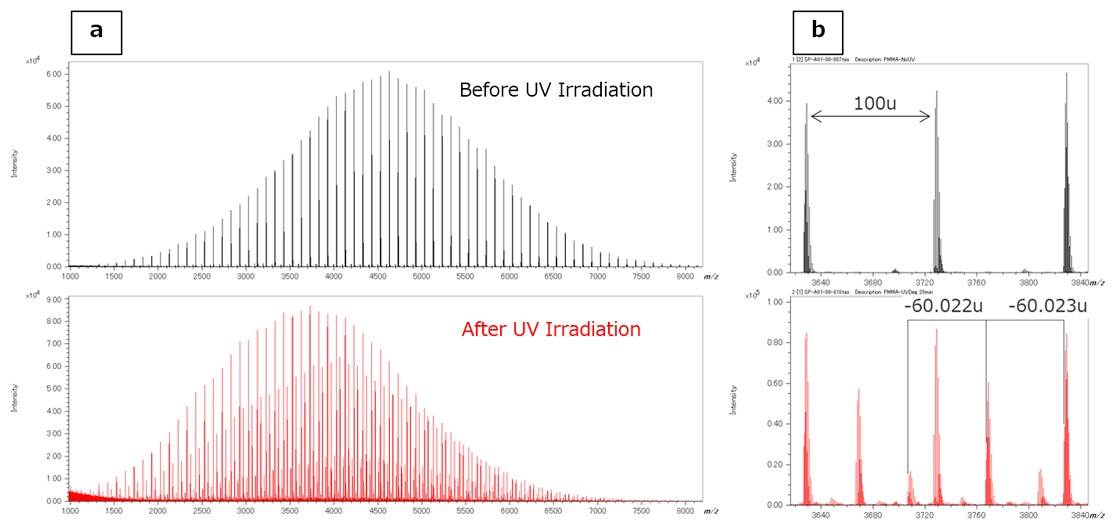
Figure 4 Mass spectra of MMA before and after UV irradiation using MALDI-TOFMS
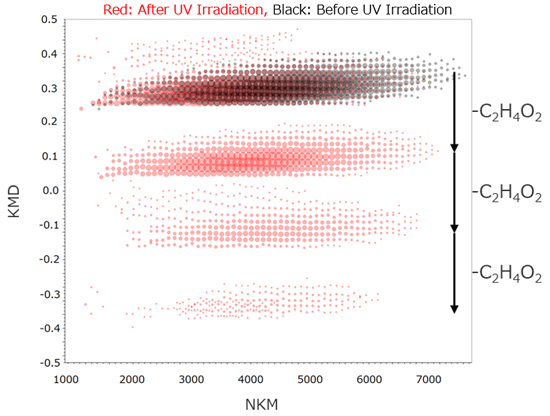
Figure 5 RKM plot of PMMA before and after UV irradiation
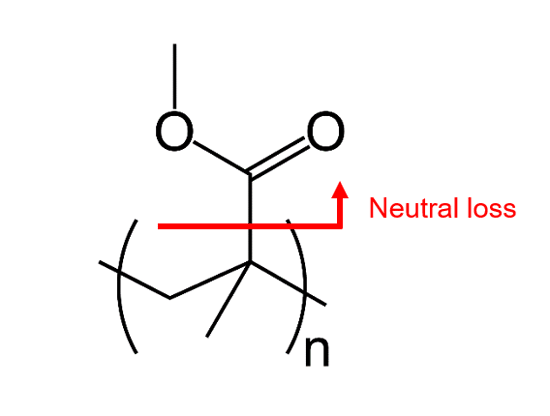
Figure 6 Estimated structure of PMMA after UV irradiation
Conclusion
The results for both Py-GC-QMS and MALDI-TOFMS showed that exposing PMMA to UV irradiation caused photodegradation of the repeating structure. High resolution MALDI-TOFMS is a valuable technique for polymer analysis because it ionizes the intact polymer for accurate mass measurements that can be used for Kendrick mass defect analysis (KMD).
On the other hand, Py-GC-QMS is useful for determining the partial structure information for polymers as well as identifying changes to their substructures during degradation. As a result, Py-GC-QMS and MALDI-TOFMS are complementary methods that can be useful for monitoring the degradation of polymer materials.
- Please see the PDF file for the additional information.
Another window opens when you click. 
PDF 805.1KB
Related Products
Are you a medical professional or personnel engaged in medical care?
No
Please be reminded that these pages are not intended to provide the general public with information about the products.


