Application of HFX NMR to Facilitate the Complete Assignment of the Anti-fungal Agent Voriconazole
AN-NM20170030 NMR application team, JEOL USA Inc.
INTRODUCTION
Fluorine is found with an ever-increasing frequency in materials science and both legal and illicit drugs [ref 1-5]. In this application note results are presented to illustrate the simplification afforded by the routine application of triple-resonance NMR to clearly assign voriconazole, a molecule containing proton, carbon, and nitrogen molecules with many atoms exhibiting J-coupling to fluorine. The ROYALPROBE HFX is a completely new probe technology utilizing magnetic coupling to afford highly efficient HF-X tuning which can function as a simple switch to highest sensitivity dedicated 1H or 19F or very well balanced dual 1H/19F performance on demand. References 6-8 detail the technological developments for the ROYALPROBE HFX.
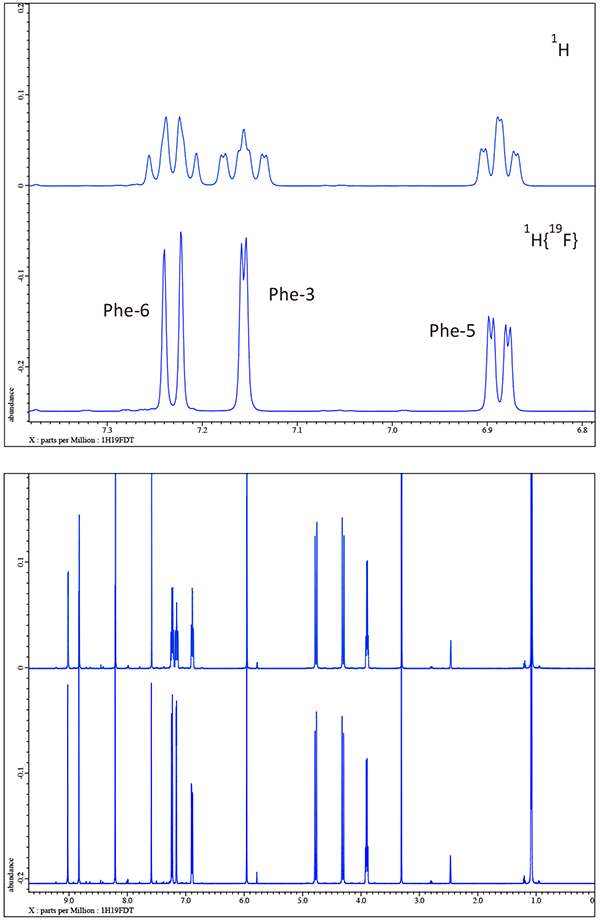
The 1H NMR spectrum acquired at 500 MHz with the ROYALPROBE HFX using the JNM-ECZR500 console is presented in figure 2. Because of the nature of the structure there is at first glance very little fine structure to facilitate clear assignments. Almost all resonances including what appear to be singlets are significantly sharpened by the application of 19F decoupling. However, the top pane of figure 2 detailing the phenyl region which contains two 19F resonances cannot be visually assigned without 19F decoupling. Decoupling 19F removes confusion with the three resonances for Phe-6, PHe-3, and Phe-5 being revealed with classically simple ortho, ortho-meta, and meta coupling patterns respectively... cannot be visually assigned without 19F decoupling.
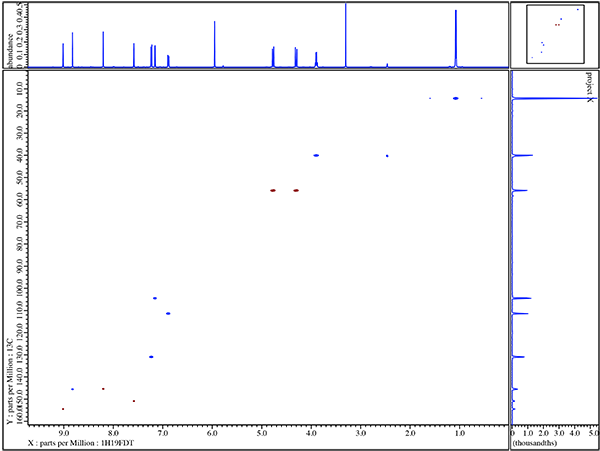
In figure 2 we see a simple illustration for how 19F can interact with the 1H spectrum but far more information is easily available to actually map out which specific 19F species are interacting with which 1H resonances. NMR pulse sequences such as HFHETCOR or HFCOSY allow H-F connectivity to be mapped in two dimensions. Figure 3 shows the HFCOSY 2D result obtained for Voriconazole.
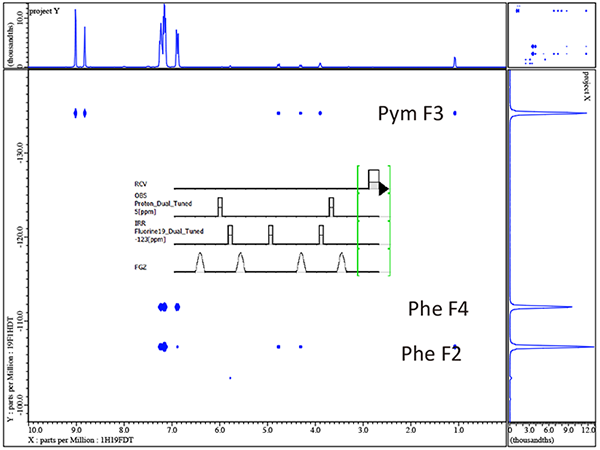
A very powerful feature in this particular example is the fact that taking the simple 1D proton results from figure 2 into account where the 3 phenyl protons are easily assigned by the 1H NMR coupling patterns the HF-COSY now allows unambiguous assignment of each 19F molecule in voriconazole. In addition the HF-COSY reveals by way of the observed responses into the 1H spectrum the singlets which are H-4 and H-6 in the pyrimidine ring. It is not yet possible to assign which singlet in the 8.8-9.0ppm region is Pym H-4 or Pym H6 so further experiments will be necessary.
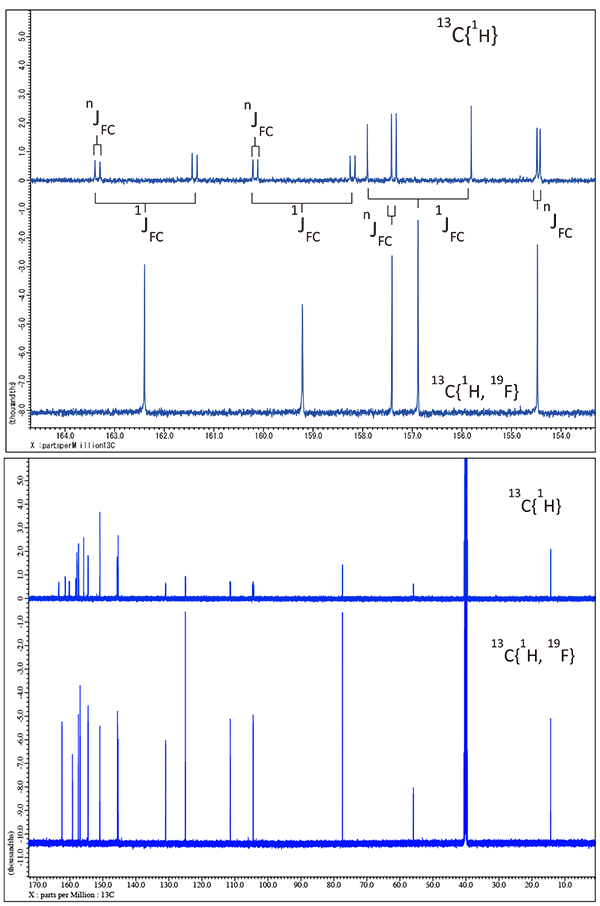
This is a good time to introduce the interactions between 19F upon the 13C NMR spectrum. In figure 4 a significant signal enhancement and simplification can be seen for the 13C spectrum brought about by the dual decoupling of 1H & 19F as opposed to only decoupling 1H. By simply comparing the 13C spectra with only 1H decoupling to a spectrum with dual {1H 19F} decoupling all carbons with attached 19F atoms now become apparent. In addition all 19F /13C coupling constants can be extract by simple inspection. Note that actual assignments cannot be made quite yet but it should be apparent that access to 1H/19F and now 13C {1H 19F } data has easily provided quite a bit of information in addition to the sensitivity enhancements.
At this point in the assignment process it is time to introduce heteronuclear experiments involve all three nuclei, 1H, 13C and 19F. While any 2 or 3 dimensional combination is possible we will focus on the higher sensitivity experiments which correlate either 1H or 19F to 13C with application of respectively either 19F or 1H decoupling in all dimensions. The 1-bond HSQC-based correlation experiments are of course purely bookkeeping in nature by allowing a simple mapping of directly connected protons or fluorines to 13C. For cases where any of the assignments of an individual species are known it allows assignment of the directly connected partner by simple inspection.
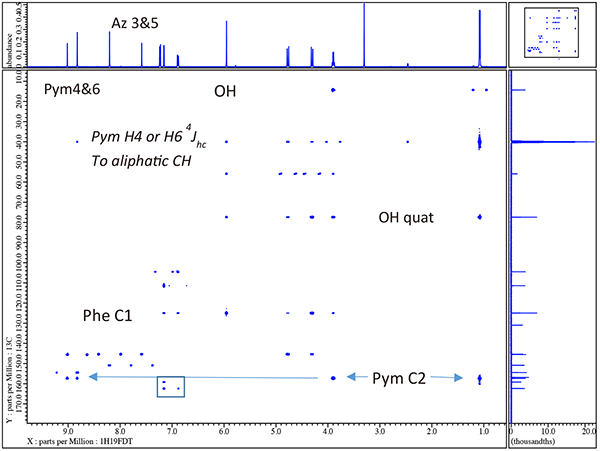
To this point we have only actually assigned the 3 fluorine atoms (via the HF-COSY and simple 19F decoupled 1H spectrum) the 3 protons in the phenyl ring. Thus, 1H/13C HSQC would provide all directly correlated H-C pairs and transfer the assignments for protons Phe6, Phe3, & Phe5 to the carbon partners. In addition the multiplicity-edited HSQC would allow confirmation of the rather obvious assignments for the OH (missing in HSQC) and the aliphatic CH, CH2 and CH3 species. The 1H/13C {19F} edited C2HSQC result for voriconazole is shown in figure 5.
Of further interest in figure 5 the lone aliphatic CH carbon is found near 40ppm which was completely obscured by the DMSOd6 solvent in the 1D carbon NMR spectrum in figure 4.
Before proceeding to the 19F/13C {1H} C2HSQC experiment it is important to provide a few details regarding the choice of C2HSQC [ref 9] for this work. C2HSQC utilizes short BIP inversion pulses for both 13C and 19F 180 degree pulses. In reference 9 the authors clearly illustrate the large increase in performance which is obtained by use of doubly compensated multiplicity- edited HSQC experiments compared to single compensation, or simple square pulse experiments. This is because the band-width required for 13C, and especially 19F, simply renders to use of simple square 180 pulses as nearly useless. The choice of BIP inversion pulses [ref 10] in tailored pairs to effect refocusing is also advantageous because of the very large 19F/13C coupling constants (260 – 280 Hz) which provides very little time for the 1/(2*(1J) required for INEPT transfer.
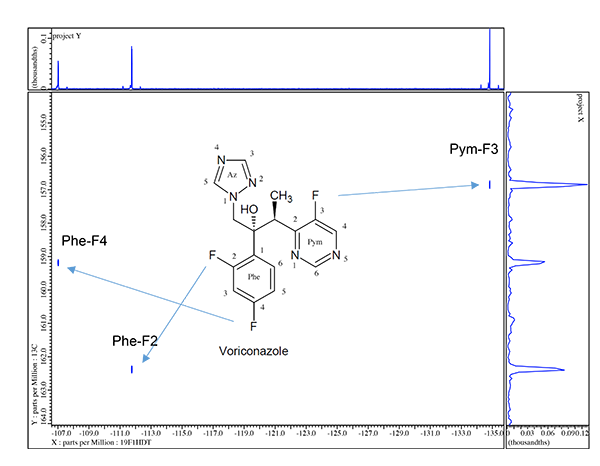
In table 1 we will list all resonance chemical shifts for 1H, 13C, and 19F and list all assignments to this point. So far as 1H, 13C and 19F are concerned everything is assigned except for the 2 CH resonances in both the triazole and pyrimidine rings and the 2 aromatic quaternary carbons. To complete the assignments as well as add the 5 15N resonances we must employ long range heteronuclear connectivity experiments.
| 13C | 1H | 19F | Multiplicity | Assign |
| 162.37 | -- | -111.76 | CF | Phe4 |
| 159.19 | -- | -107.02 | CF | Phe2 |
| 157.37 | -- | -- | quat | ? |
| 156.86 | -- | -134.83 | CF | Pym3 |
| 154.46 | 9.015 | -- | CH | ? |
| 150.87 | 7.585 | -- | CH | ? |
| 145.55 | 8.824 | -- | CH | ? |
| 145.36 | 8.202 | -- | CH | ? |
| 130.96 | 7.231 | -- | CH | Phe6 |
| 124.97 | -- | -- | quat | ? |
| 111.38 | 6.892 | -- | CH | Phe5 |
| 104.46 | 7.157 | -- | CH | Phe3 |
| 77.4 | -- | -- | quat | C-OH |
| 55.89 | 4.774;4.304 | -- | CH2 | CH2 at triazole N1 |
| 40.06 | 3.899 | -- | CH | CH-with CH3 |
| 14.32 | 1.079 | -- | CH3 | CH3 |
| -- | 5.95 | -- | OH | OH |
The gHMBCAD pulse sequence [ref 11] was used to acquire 1H/13C {19F} and 19F/13C {1H} long-range correlation experiments to complete the missing 1H & 13C assignments. In addition 1H/15N {19F} and 19F/15N {1H} data was acquired to assign the 5 nitrogens in voriconazole.
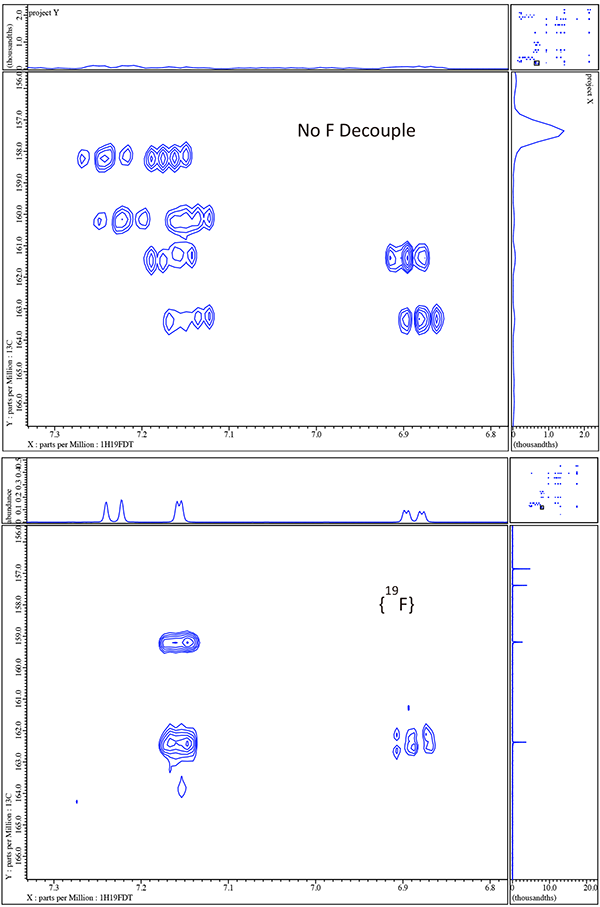
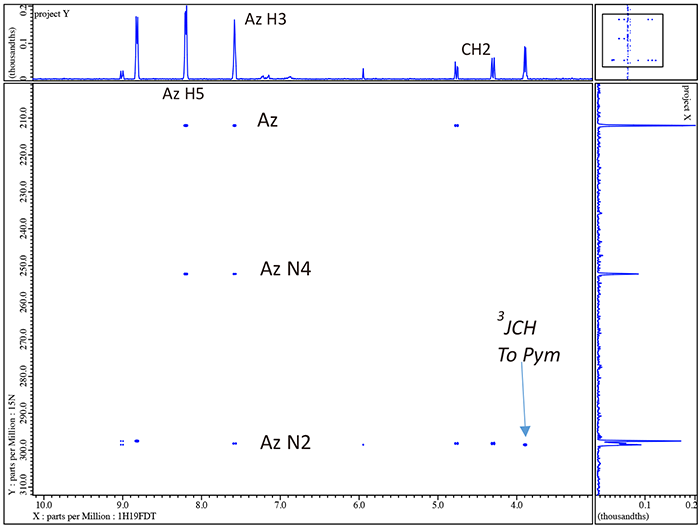
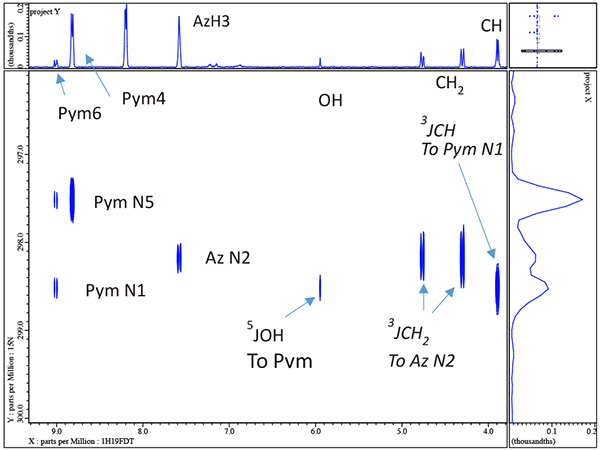
Note that with just a quick inspection of the gHMBCAD in figure 7 we can now assign the quaternary carbons Pym2 and Ph1 as well as the rather obvious aliphatic quaternary carbon to which the OH is attached. Furthermore we also now clearly know which of the 4 aromatic singlets are associated with both the triazole and pyrimidine rings though we cannot yet say for the case of the pyrimidine which is H4 and which is H6 or for the trizole which is H3 and which is H5. Thus other than the ambiguity for Pym 4 & 6 and Az 3 & 5 we have completely and easily assigned voriconazole. Before we resolve these ambiguities by 1H/15N gHMBCAD we will explore the effects of decoupling 19F in 1H/13C gHMBCAD.
In figure 8 we compare 1H/13C gHMBCAD for the small boxed region in figure 7. Note the large simplification resulting from decoupling {19F} in both dimensions. Now it is time to move to 1H/15N gHMBCAD in figures 9 & 10 and see how easily we can resolve the ambiguity for the 2 pairs of singlets in the pyrimidine and triazole rings.
| 13C | 1H | 19F | Multiplicity | Assign |
 |
| 162.37 | -- | -111.76 | CF | Phe4 | |
| 159.19 | -- | -107.02 | CF | Phe2 | |
| 157.37 | -- | -- | quat | Pym2 | |
| 156.86 | -- | -134.83 | CF | Pym3 | |
| 154.46 | 9.015 | -- | CH | Pym4 | |
| 150.87 | 7.585 | -- | CH | Az3 | |
| 145.55 | 8.824 | -- | CH | Pym6 | |
| 145.36 | 8.202 | -- | CH | Az5 | |
| 130.96 | 7.231 | -- | CH | Phe6 | |
| 124.97 | -- | -- | quat | Phe1 | |
| 111.38 | 6.892 | -- | CH | Phe5 | |
| 104.46 | 7.157 | -- | CH | Phe3 | |
| 77.4 | -- | -- | quat | C-OH | |
| 55.89 | 4.774;4.304 | -- | CH2 | CH2 at triazole N1 | |
| 40.06 | 3.899 | -- | CH | CH-with CH3 | |
| 14.32 | 1.079 | -- | CH3 | CH3 | |
| -- | 5.95 | -- | OH | OH | |
| 15N | |||||
| 299.91 | AZ2 | ||||
| 298.52 | Pym1 | ||||
| 297.52 | Pym5 | ||||
| 252.18 | Az4 | ||||
| 211.99 | Az1 |
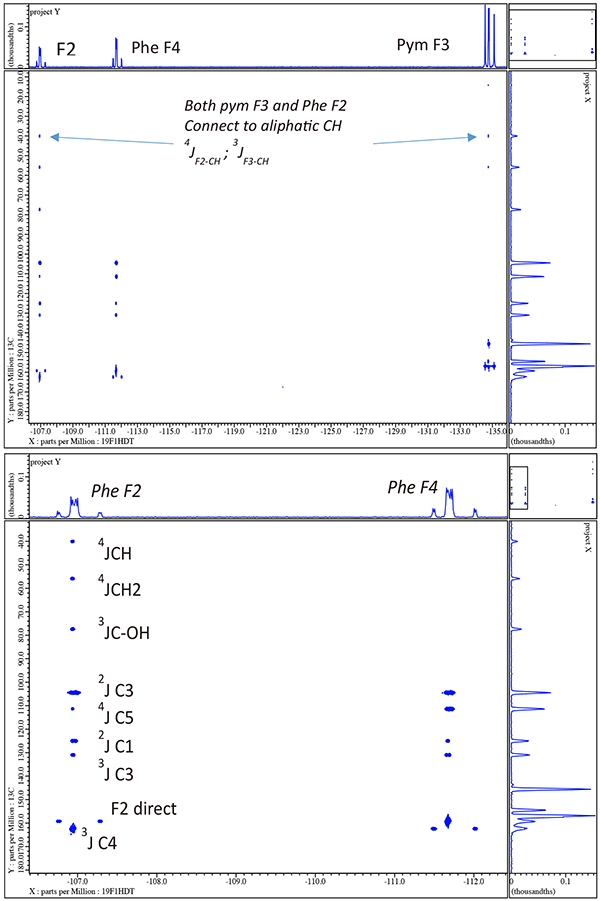
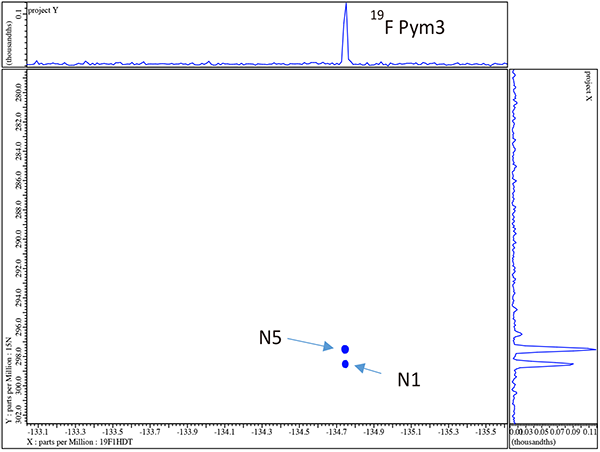
At this point we have achieved the goal of completely assigning all resonances for the voriconazole sample but have the luxury being able to now employ 19F/13C and 19F/15N gHMBCAD with full {1H} decoupling as a completely independent check of many of the assignments. The 19F/13C gHMBC is shown in figure 11 and the 19F/15N gHMBCAD in figure 12.
CONCLUSION
By using HF-X NMR in a variety of one and two-dimensional experiments it is possible to clearly assign all resonances in an important drug such as the chosen example of voriconazole. The ROYALPROBE HFX allows easy on-demand access to such an array of experiments in a full automation environment.
REFERENCES
- Wang et al, Chem. Rev. 2014, 114, p2432−2506
- Zhou et al, Chem. Rev. 2016, 116, p422−518
- http://www.slweb.org/ftrcfluorinatedpharm.html
- Banister et al, ACS Chem. Neurosci., 2015, 6 (8), p1445–1458
- Trachsel, D., Drug Test. Analysis, 2012, 4: p577–590
- B. Taber and A.P. Zens, J. Magn. Reson., 259, 2015, p114-120
- P. Bowyer, J. Finnigan, B. Marsden, B. Taber, and A. Zens, J. Magn. Reson., 2015, p190-1
- V. Lim, B. Marsden, B. Taber, and A. Zens, J. Magn. Reson., 2016, p.268
- H. Hu, K. Krishnamurthy, Magn. Res. Chem., 46 issue 7, 2008, p 683-689
- M. Smith, H. Hu, and A.J. Shaka, J. Magn. Reson., 151, 2001, p 269-283
- R. Crouch, R. Boyer, R. Johnson, and K. Krishnamurthy, Magn. Reson. Chem., 42, 2004, p301-307
Related Products
Are you a medical professional or personnel engaged in medical care?
No
Please be reminded that these pages are not intended to provide the general public with information about the products.
