Biological Electron Microscopy; Seeking Evermore Significant Data for New Discoveries in Biological Research

INTERVIEW 08
Roland Fleck
Professor at King’s College London, United Kingdom
Understanding cellular and molecular structures is a principal foundation of biomedical research. Professor Roland Fleck, who is the Director of CUI (Centre for Ultrastructural Imaging) at King’s College London, dedicates himself to high resolution imaging of their structures using JEOL electron microscopes. He uses both scanning and transmission electron microscopes, e.g., JSM-7800FPRIME, JEM-F200 well as other JEOL instruments in partnership with JEOL. The centre, which is a JEOL Centre for Advanced Technology (JCAT) specializes in both room temperature and cryo electron microscopy techniques.
Why it is important to understand complex biological process.
CUI (Centre for Ultrastructural Imaging) at King’s College London, under the directorship of Professor Fleck focuses on the application of advanced imaging techniques using both the transmission electron microscope (TEM) and the scanning electron microscope (SEM) to understand complex biological process. “Biological process” covers various kinds of biological phenomena, including genomic organization, signal transduction, cell cycle progression, and other complex systems.
The human body has different areas specialized for different functions: eyes see, legs walk. Spatial specialization of function occurs at every scale: within the eye, lens and photoreceptor cells contribute differently to vision, and different molecules clustered into different parts of each photoreceptor cell perform different tasks (some detect light, others let the brain know that light is detected). However, the molecular level of magnification is around 1,000,000x. This cannot be achieved by normal light microscopes, and requires microscopes using the shorter wavelength of electrons. However, electron microscopes are normally limited to looking at very thin samples of tissue, 1/1000th the thickness of human hair, and it takes hundreds of these thin sections to contain all of one cell.
Cells and tissues must also first be prepared in for study in the electron microscope. How this preparation is done requires expertise in many different fields and research to understand and optimize specimen preparation procedures. Professor Fleck seeks to improve sample preparation through research, in order to obtain higher quality information from the sample. “It is in this context that the collaborations with JEOL are so valuable,” says Professor Fleck.
Cryopreperation of biological tissues.
Biological tissues consist of groups of cells. For example, the heart is an organ which is composed of various tissues (myocardium, blood vessels, nerves, etc.) and plays a biological role with these tissues being bound under a certain order. So, understanding the morphologies and structures of tissues is essential.
Cryo-electron microscopy enables observation of such a tissue sample by using low temperatures to preserve its “native” state. The sample is cooled and kept at very low temperatures in a “cryo” stage fitted to the electron microscope. The avoidance of crystalline ice upon freezing (vitrification) is fundamental to unlocking the resolving potential of cryo-electron microscopy. Vitrification allows for structures to be viewed in their native state, avoiding the artefacts commonly associated with chemical fixation and dehydration preparation techniques as well as the ultrastructural disruption attributed to intracellular ice formation.
Professor Fleck uses many cryo techniques which each address the challenge of avoiding ice crystals on freezing. Rapid cooling allows vitrification of small volumes through the use of an intermediate cryogen (e.g., ethane or propane). This avoids the Leidenfrost effect (where a film of insulating vapour, created by raising the cryogen above its boiling point, slows down cooling). This is the approach established by Jacques Dubochet (Nobel Prize Chemistry in 2017) to vitrify small isolated particles as a film on a TEM grid for single particle TEM. For thicker samples of whole cells or tissues a high pressure freezer is used. The high pressure freezing (HPF) technique is based on the physics of water: pressure of 2100 bars applied during freezing acts as a physical cryoprotectant; high pressure counteracts the expansion of water upon crystallization, thus slows down ice crystal formation and reduces the critical cooling-rate required for vitrification (10,000°C.s-1).
Vitrified tissue blocks can be processed by freeze substitution, sectioned at cryogenic temperatures (CEMOVIS) or fractured and directly observed in the cryo SEM or following the generation of replica viewed in the TEM. Professor Fleck employs each of these techniques to address different questions. For example HPF is now integrated with electrical stimulation and light stimulation allowing high levels of temporal correlation between stimulation and immobilization when applied to study how information is transferred from one neuron to another in the brain.
In light microscopy fluorescent tags are used to display proteins confined to compartments in living cells, but give no glimpse of the underlying ultrastructure. The electron microscope has the resolution to display cellular ultrastructure but cannot “see” the fluorescent tags. Instead immunogold labelling is used to visualize (tag) the locations (localization) of target (specific) cells and/or tissues with an electron microscope, by utilizing antigen-antibody reactions where antibodies bind specifically to antigenic cells and/or tissues. For this technique, a frozen ultrathin sectioning method (Tokuyasu technique) firstly introduces a cryoprotectant (sucrose) to suppress ice crystals formation during comparatively slow cooling rates achieved by plunging a sample directly into liquid nitrogen (~ <100°C.s-1). The tissue is then sectioned by cryo ultramicrotomy and labelled with immunogold.
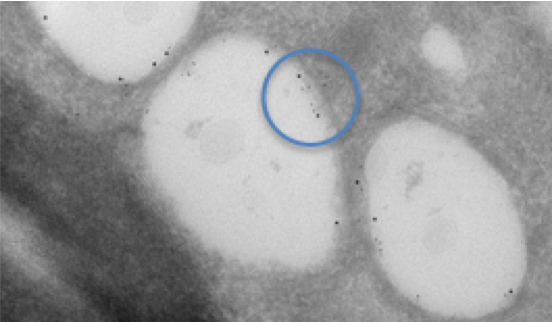
Fig. 1. Dual labelling of lipid droplets in a cell infected with Hepatitis C virus. Two different sizes of colloidal gold are used to label different proteins of interest and show that they are each localised on the inner membrane of the lipid droplet. The tissue was prepared, sectioned and labelled using the Tokuyasu technique. Dual labelling is highlighted in the blue circle.
Importance of Understanding Cellular and Molecular Structures by SEM and TEM to Parasitology.
Understanding the cellular and molecular structures of Malaria is essential to develop effective anti-malarial treatments. Malaria, Plasmodium falciparum, parasites develop within red blood cells (erythrocytes) and are contained inside a membrane-enclosed parasitophorous vacuole. During their development the parasite modifies the surface of the red blood cell leading to rigidification of the cytoskeleton in infected cells, and for the transmission to the cytoskeleton of shear forces experienced by adhering cells. Once fully developed a multistage process termed egress occurs. To do this, the parasites orchestrate a highly regulated sequence of membrane permeabilization and breakage steps culminating in the explosive release of parasites for a new round of infection.
Understanding these processes is a key to unlocking the life cycle of malaria. For this study, electron tomography is useful. Electron tomography is a method to reconstruct three-dimensional internal structures of a sample through computer image processing of many projection images, which are acquired from sequential tilt-series images of the sample. Professor Fleck uses electron tomography of high pressure frozen, freeze substituted infected red blood cells and cryo electron tomography of vitrified red blood cell ghosts to examine the 3-dimensional modification of the red blood cell surface. A highly organized knob skeleton composed of a spiral structure coated by an electron-dense layer is underlying the knob membrane. This knob skeleton is connected by multiple links to the erythrocyte cytoskeleton. The arrangement of membrane proteins in the knobs was then visualized by high-resolution freeze-fracture SEM. Samples were prepared by high pressure freezing and freeze fracture of live parasites. Membrane structures were revealed at high resolution and knob structures were distinct from that in the surrounding erythrocyte membrane, with a structure at the apex that likely represents the adhesion site. Thus, it was shown that erythrocyte knobs in P. falciparum infection contain a highly organized skeleton structure underlying a specialized region of membrane.
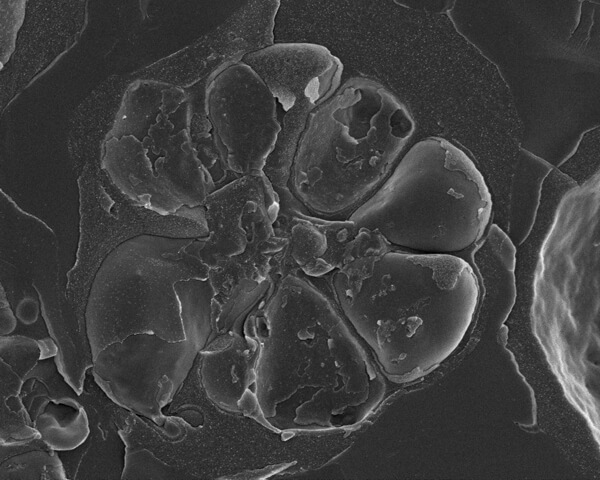
Fig. 2. Malaria, Plasmodium falciparum, parasites developing within a red blood cell (erythrocytes). The seven merozoites are contained within a membrane-enclosed parasitophorous vacuole. The merozoites are located around a central residual body. Due to the fracture process many individual membranes are revealed.
3D Reconstruction using Serial Block Face SEM
Studying the brain is crucial because revealing the connections of nerve cells in a brain leads to an understanding of mechanisms of learning and memory. Connectome analysis aiming to reveal all connections among all nerve cells in a brain is the objective. For visualizing these connections a TEM allows for observation of the connections between neurons, but the observable area is very thin and limited. “So in this case, Serial Block Face SEM (SBF-SEM) is very effective,” Professor Fleck points out.
Serial Block Face SEM (SBF-SEM) is a method to reconstruct the three-dimensional (3D) structure of a specimen block using a SEM. This method is mainly used for 3D reconstruction of soft materials such as resin-embedded neuronal “brain” tissue. The block specimen of the brain tissue is sliced with a thickness of ~30 nm to a volume of >500 μm from the surface with an ultramicrotome incorporated in the SEM specimen chamber, and then each face exposed by slicing is observed. Collected images are stacked to obtain the 3D structure of the specimen block.
Charging of samples during SBF-SEM can limit the quality of data generated to improve conductivity of the tissue block, so an OTO technique is used (osmium tetroxide–thiocarbohydrazide–osmium tetroxide). This protocol enhances the contrast of lipids, long pre-dating SBF-SEM. Professor Fleck uses SBF-SEM and adapts and modifies these older processing techniques to produce even staining throughout of the tissue block, without the danger of large precipitates to give a strong backscatter signal and high conductivity in the SBF-SEM. Using this technique, Professor Fleck mapped the distribution of the structure of presynaptic terminals in CA1 stratum oriens, a region of the hippocampus that receives ordered axonal inputs. Data stacks from multiple regions were reconstructed and, by using reference markers in the tissue, the data correlated to synaptic inputs determined by collaborators. Although dendritic segments showed a wide distribution in the size of excitatory inputs, there was a strong correlation between morphological measures of presynaptic and postsynaptic compartments. Presynaptic boutons that form onto basal dendrites of CA1 pyramidal neurons display a decrease in active zone (AZ) size with distance from the soma. This provides a distance-dependent increase in short-term facilitation (function). Thus, the spatial distribution of short-term facilitation modulated by morphology serves to compensate for the electrotonic attenuation of subthreshold distal inputs during repeated stimulation and fine-tunes the preferred input frequency of dendritic domains.
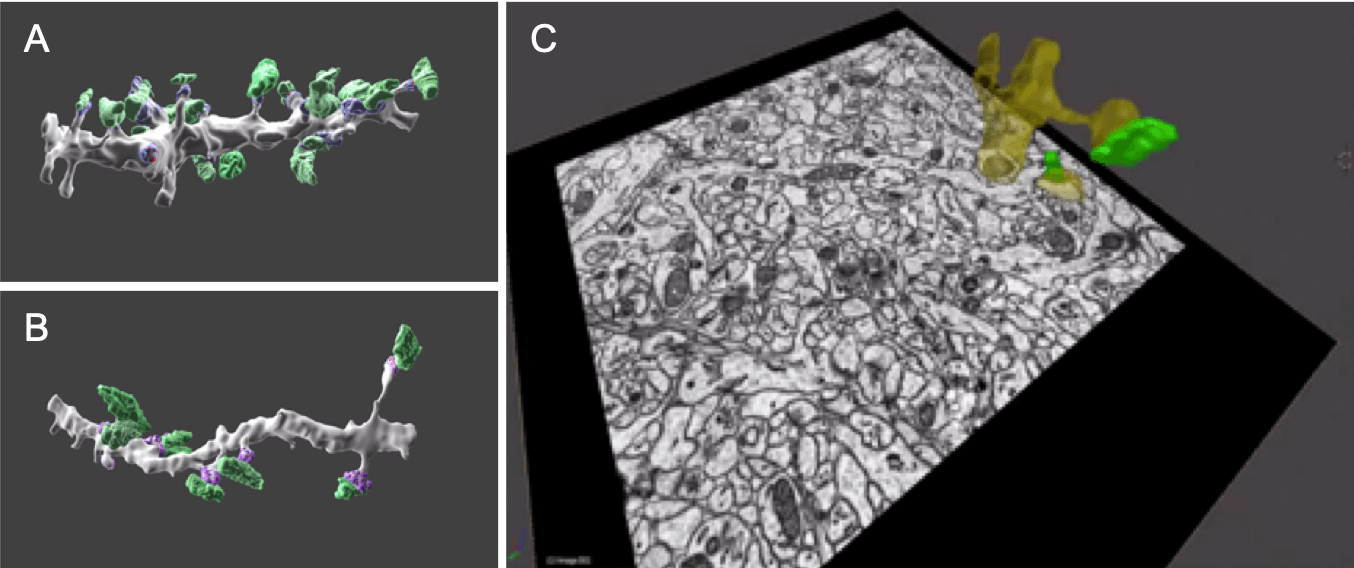
Fig. 3. 3D-View: Serial block-face scanning electron microscopy of the hippocampus which contains many neurons. Plate A and B show presynaptic boutons coloured green and post synaptic densities purple, A is from the proximal and B the distal area of the dendrite. Plate C shows a single slice from a serial block face with the reconstructed dendritic compartment (yellow) and presynaptic bouton (green) superimposed on the image. By collecting many individual slices, it is possible to reconstruct the tissue in 3D.
Correlative Light and Electron Microscopy (CLEM) for combining optical microscope and electron microscope
This method (CLEM) employs a unique combination of features of the optical microscope and the electron microscope. It is used to firstly identify proteins or tissues using a fluorescence microscope where the proteins or tissues are subjected to fluorescent staining (labelling), and then to observe the morphology or structure of the identified target regions using an electron microscope with high spatial resolution. CLEM is widely used in bio-science, by which analysis of functions and fine morphology (structure) of labelled proteins or tissues can be made. CLEM can be combined with either TEM or SEM electron microscopes and with the addition of specialist cryogenic stages can support correlation between fluorescence and high resolution cryo EM instruments. In CUI there are multiple CLEM workflows to follow. These include a Nikon to JEOL SEM room temperature workflow where cells are grown on a special tissue culture dish which supports live cell imaging. These tissue culture dishes enable correlation between a cell process and imaging in the SEM after room temperature sample processing.
JEOL Centre for Advanced Technology for Further Advancement of Life Science
Combining cryo electron microscopy and structural biology approaches to determine 3D cellular structures “in situ” from sections of vitrified cells and tissues is an emerging frontier for cell biology. It requires not only a cryo TEM but also cryo fixation and cryo sectioning tools. This includes optimising the latest focused ion beam SEM (FIB-SEM) from a JEOL JIB-4700F to harness FIB vitreous lamella preparation and contamination free transfer to the cryo TEM.
Professor Fleck says, “Biological Electron Microscopy has made remarkable progress in this last decade. We will now seek to explore new challenges through a close collaboration with JEOL. We have many, many ideas to exchange and proposals for cutting-edge instrument development. I am excited to be part of these future innovations.”
JEOL Centre for Advanced Technology (JCAT) also plays an important role that contributes to the society. JCAT is providing various technologies to the local hospitals for diagnosis of kidney disease, skin disease, etc. the JEOL RUBY camera equipped on JEOL JEM-1400Plus is very effective for conducting biopsy with easy operation.
Professor Flecks emphasizes “JCAT, established based on a partnership with JEOL, will become a crucial platform where new ideas of instruments and techniques will be born owing to our constructive cooperative relationships, leading to further advancement of lifescience study.”

Roland Fleck
Professor at King’s College London, United Kingdom
Professor Fleck studied Applied Marine Biology at Heriot Watt University in Edinburgh, Scotland before moving to the Institute for Freshwater Biology to study for his PhD in “Mechanisms of cell damage and recovery in cryo preserved freshwater protists”. It was during his PhD that he first started working with electron microscopes, a JEOL JEM-100CX. After his PhD he moved to Cornell University to research cold acclimation in plants. During his time at Cornell University he worked extensively with the Freeze Fracture/ Freeze Etch replica technique. He returned to the United Kingdom to join the National Institute for Biological Standards and Control where he worked to develop and standardize functional cell-based assays as well as leading and developing their electron microscopy and cryo-imaging facility. He left NIBSC to assume the role of Director, of the Centre for Ultrastructural Imaging at King’s College London.
Posted:March 2019
